Team:TU Munich/Modeling/Priors
From 2012.igem.org
(→Yeast mRNA Degradation Rate) |
|||
| Line 1: | Line 1: | ||
{{Team:TU_Munich/Header}} | {{Team:TU_Munich/Header}} | ||
| - | |||
= Prior Data = | = Prior Data = | ||
<hr/> | <hr/> | ||
Revision as of 16:46, 11 September 2012



Contents |
Prior Data
Yeast mRNA Degradation Rate
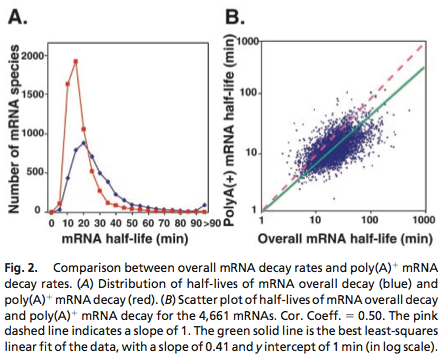
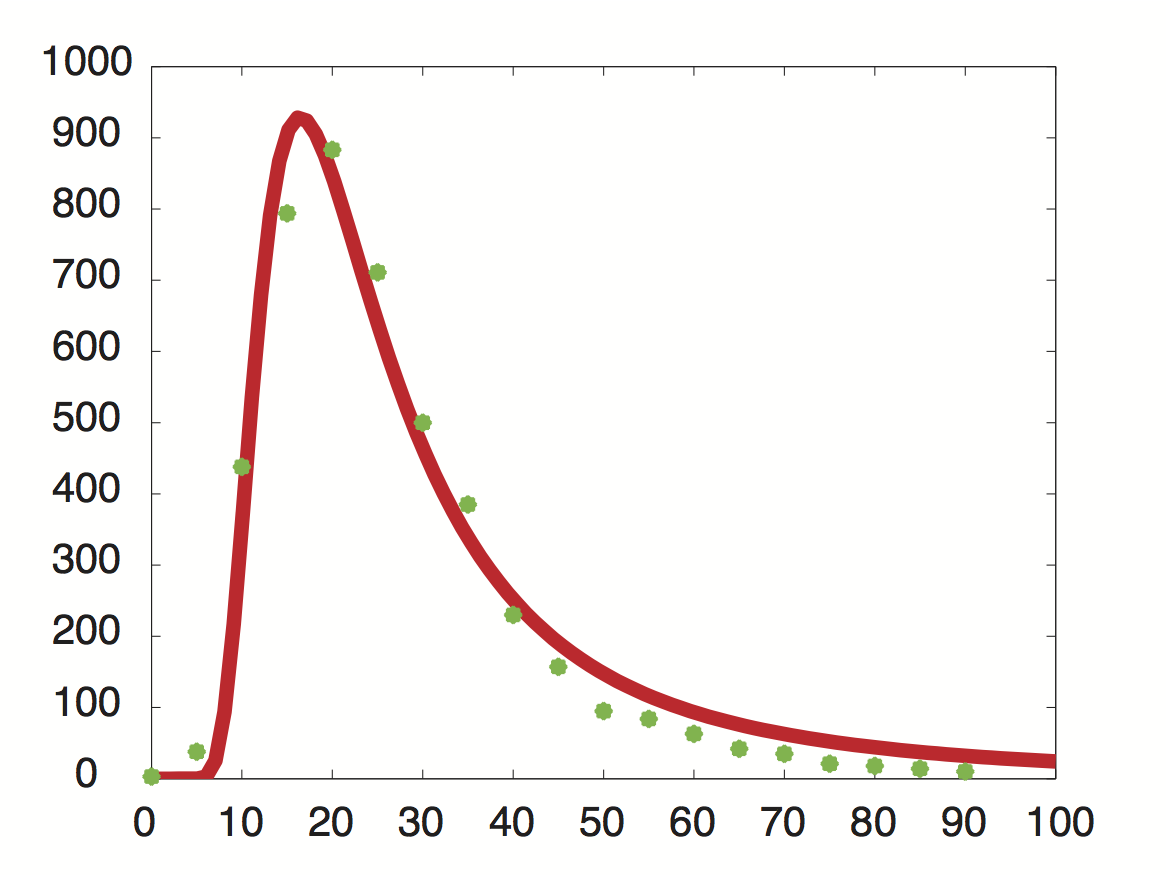
Data was obtained from the Paper (Wang et al. 2001 [http://www.pnas.org/content/99/9/5860.long]) and processed by [http://arohatgi.info/WebPlotDigitizer/app/] to obtain raw data. Using a least-squared error approximation the distribution of the half life time in was approximated as noncentral t-distribution with parameters μ = 1.769 and ν = 20.59.
dataGraph = [ 0.0018691649126431735,0.0016851538590669062 0.05978099456360327,0.01885629059542104 0.11548146330755026,0.21910551258377348 0.17122389948476902,0.396902157771723 0.2253457470848775,0.4417136917136917 0.2815821076690642,0.3552607791738227 0.3359848142456839,0.249812760682326 0.39216629434020744,0.19272091011221448 0.4465173486912618,0.11490683229813668 0.5026600896166115,0.07854043723608946 0.5569239808370243,0.04735863431515607 0.6111394480959699,0.04208365077930302 0.667233765059852,0.031624075102336016 0.7233280820237343,0.021164499425369035 0.777540321018582,0.017616637181854626 0.8373665112795547,0.010604847561369285 0.8897063570976615,0.008787334874291503 0.9420462029157682,0.006969822187213598 0.9999935434718044,0.005142624707842157 ]; X = round(dataGraph(:,1)*90); y = round(dataGraph(:,2)*2000); k(1) = 1.769292045467269; k(2) = 20.589996419308118; k(3) = 24852.48237036381; k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k);
Yeast Protein Degradation Rate
For the Degradation Rate the N-end rule (Varshavsky 1997 [http://openwetware.org/images/c/c2/The_N-end_Rule_Pathway_of_Protein_Degradation.pdf]) served as approximation for the half life time. It states that the half life time in S. cerevisiae can be approximated based on the amino acid after the initial start codon.
| Residue ! Half-life | |
|---|---|
| Arg | 2 min |
| Lys, Phe, Leu, Trp, His, Asp, Asn | 3 min |
| Tyr, Gln | 10 min |
| Ile, Glu | 30 min |
| Pro | > 5 h |
| Cys, Ala, Ser, Thr, Gly, Val, Met | > 30 h |
As these values do not give enough information to infer a proper distribution, only the two lower bounds 5 h and 30 h will serve as approximate lower bounds for the optimization routines.
Yeast Transcription Rate Rate
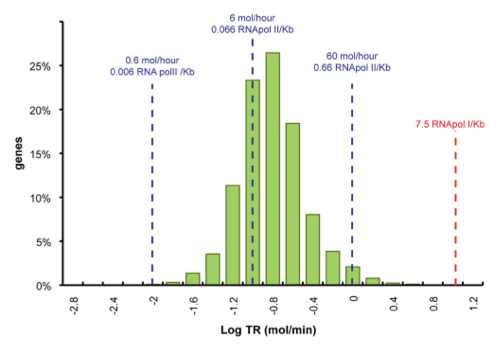
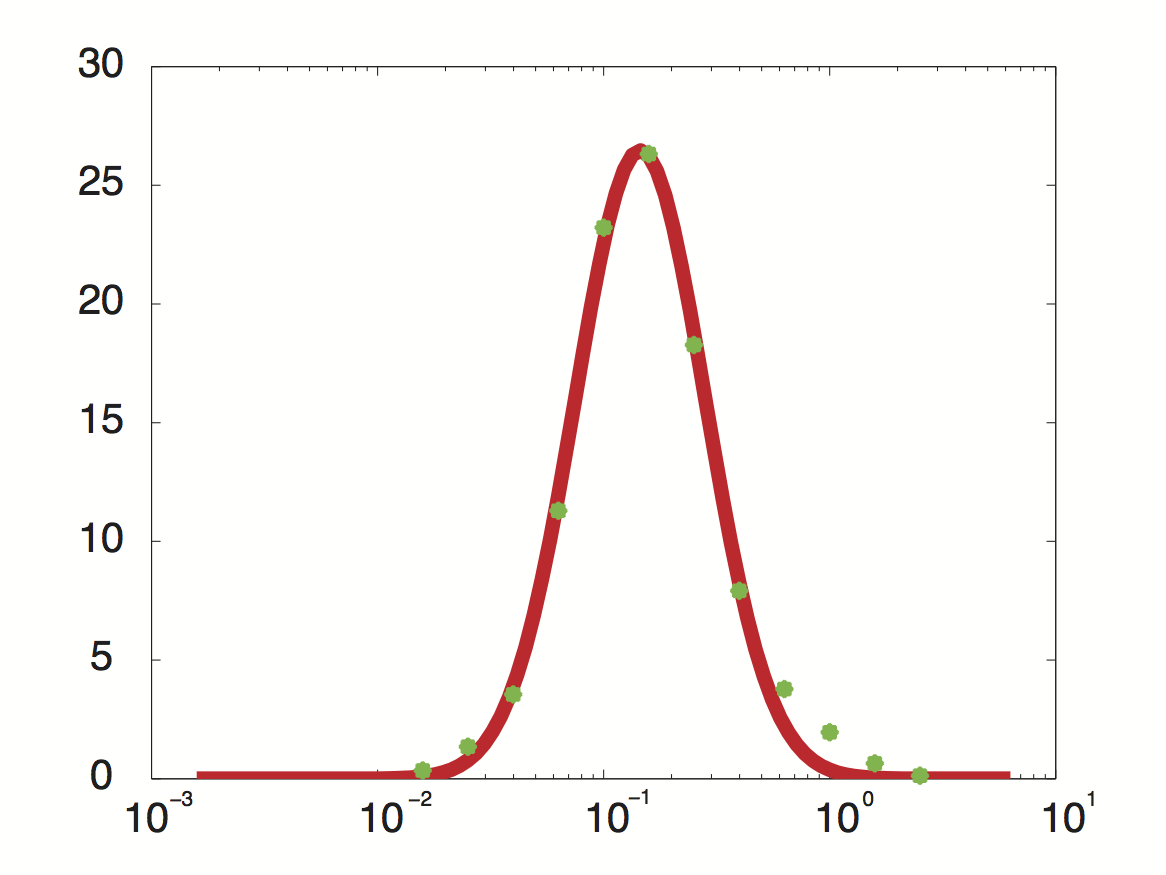
Data was obtained from the Paper (Pelechano et al. 2010 [http://www.pnas.org/content/99/9/5860.long]) and processed by [http://arohatgi.info/WebPlotDigitizer/app/] to obtain raw data. Using a least-squared error approximation the distribution of the transcription rate was approximated as log-normal distribution' with parameters μ = -1.492 and σ = 0.661;.
dataGraph = [ -1.8,0.3442950751957339 -1.6,1.3525375039897853 -1.4,3.5492668181220783 -1.2,11.28874786429094 -1.0,23.213749272450762 -0.8,26.31522126884587 -0.6,18.273455248681024 -0.4,7.913623476840467 -0.2,3.7755111620134825 0,1.9559339854677913 0.2,0.6458759692833385 0.4,0.12767315671880167 ]; x = 10.^dataGraph(:,1); y = dataGraph(:,2); k(1) = -0.8; k(2) = 0.2; k(3) = 25; k=fminunc(@(z) sum((y-z(3)*lognpdf(x,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*lognpdf(x,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*lognpdf(x,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*lognpdf(x,z(1),z(2))).^2),k);
 "
"