Team:Amsterdam/software/designer/setup
From 2012.igem.org




Cellular Logbook Construct Designer: Setup
We have designed an online tool where a user can create their Cellular Logbook in silico by giving a backbone sequence and selecting one or more sensors manually or predefined from the parts registry database. An algorithm then computes one or more plasmids, taking into account the rules we use for our real life Cellular Logbook, and outputs the appropriate genbank files as well as the visualized version of the plasmid. The tool will also generate all possible permutations of gel bands which can be used as a read-out sheet for the gel electrophoresis results. By using this tool we are able to quickly design a plasmid with multiple sensors.
Contents |
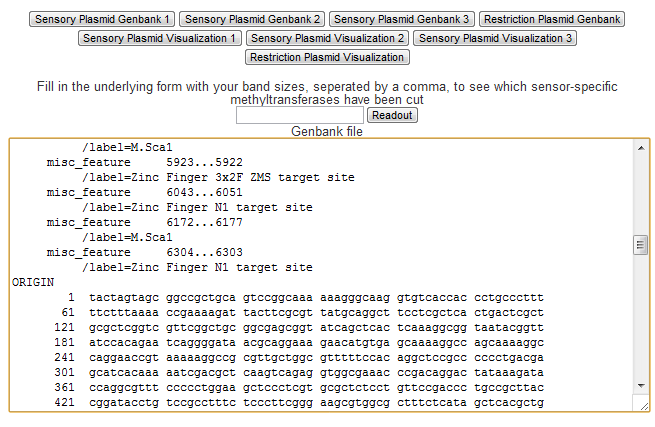
Genbank Output
Standard formats for biological data are essential to communicate and pass along biological findings. The Genbank format is the most widely used format for genome and sequence annotation. As such the Plasmid Designer is able to produce Genbank files for each plasmid with the click of a button. This way the user can use the designed construct in other biological tools such as the freeware plasmid editor ApE[http://biologylabs.utah.edu/jorgensen/wayned/ape/].
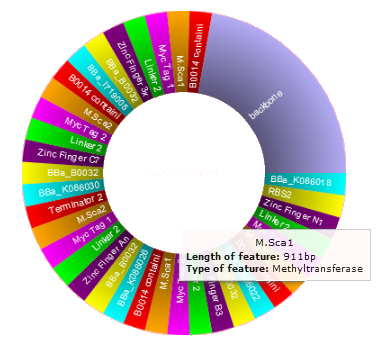
Plasmid Visualization
In order to grant the user a quick and dynamic overview of the plasmids that were just designed, a visualization of each plasmid is available with just a click on a button. Each of these visualized plasmids shows the specific features of that plasmid by color code. Hovering over each feature gives additional information, and clicking on a feature gives its DNA sequence.
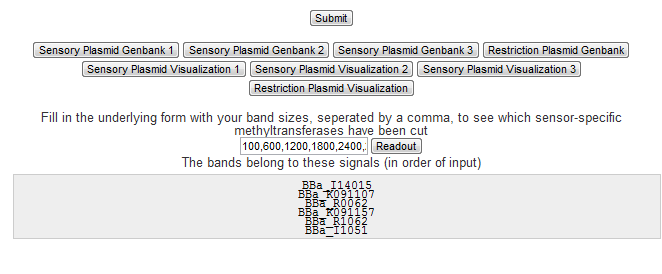
Gel Readout
The amount of gel bands increases exponentially as more sensors are engineered into the construct. Therefore having to figure out which signals have come to expression and which have not can prove to be an impossible task. We have included an algorithm inside the tool that computes all possible band combinations, and selects the bands that belong to each band position given by the user.
 "
"