From 2012.igem.org

Team:Technion/11_September_2012
Ilya
- Minipreped the picked colonies for testing the clones of Fus2 and Fus2del in pCP.
- Did a restriciton digest with BamHI-HF to test the colonies for the correct insert. 3/3 clones of Fus2del and 3/4 clones of Fus2 were positive. I picked one of each of the clones for plate reader tomorrow.
- Put starters for plate reader tomorrow.
- Tested ladders on a 0.6% agarose gel. Ran for an hour and 10 mins at 40V on a minigel. The 1Kb opened but the high range (HR) ladder didn't open. We will use a 0.4% agarose gel in the future attempts.
- Hila and I did the first attempt for gibson on the phage fragments. We attempted an assembly of fragments #4 (6150bp) and #5 (8080bp). The results are presented in the following gel pic:
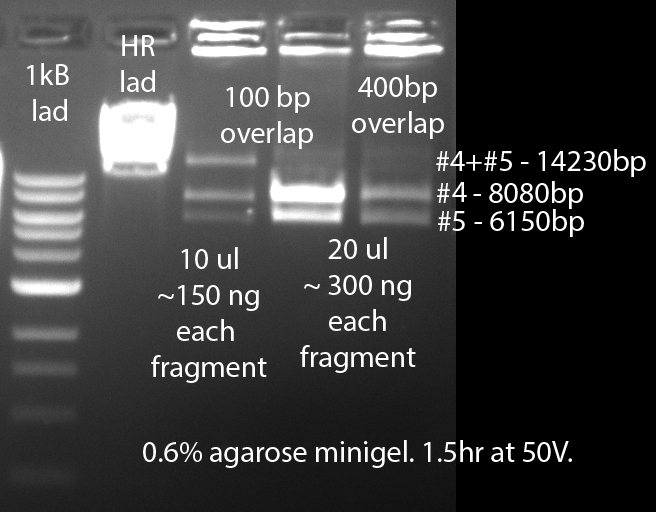
As you can see in the attached file, only one of our three attempts today actually worked. The reaction conditions for the attempt that worked were: 100bp overlaps, ~150ng DNA of each of the templates (~0.025-0.03 pmoles), 5ul of Gibson mix and 5 ul of DNA fragments (10ul final volume). All reactions were incubated for 1 hr at 50 degrees as instructed by the manual.
I think that the 20 ul final volume reaction with the 100bp didn't work because there was too much DNA. The 400bp overlap might still work with additional exonuclease and in a 10ul volume with ~150ng of each fragment.
Inbal
-Miniprep for two samples out of three from the starters I made yesterday- because one of them has been contaminated. The conc's are: 146.5 and 142 ng/ul. I didn't know for sure if the insert (eT7) is within the plasmid (pSB1AK3) so I digested the plasmid with EcoRI. After running the products on a gel, comparison to the uncut plasmid, I got the desired band- 6kb approximately! The clone works!
- Shachar helped me to do a second transformation of native T3 to pSB1AK3 (2 transformations of 10ul and 6 ul from the ligation), we plated on 4 amp resistance plates (100ul and rest) and incubate them overnight at 37C.
- restriction of eT7+pSB1AK3 with XbaI, then I cleaned the products from buffers,etc.. and continue it to a cip reaction, after that I measured the concentration: 13.2 ng/ul. I ligated an inducible promoter (pTetO) attached to a reporter gene (m-cherry) to eT7+pSB1AK3, 37C for 1 hour.
- I transformed the ligation product into TOP10 component cells +control of cip.
- Starter for eT7+pSB1AK3 for cloning it to shipping plasmid.
Asaf
I cleaned the DNA from the extracted gel. I got a very low DNA concentration.
Hila
- Gel purification for F4 (400bp overlap), F4*(100bp overlap) and F5.
- Colony PCR for additional five colonies of fragments 4C, 5C and 7C. I found one positive colony for fragment 5.
- Transformation of ligated 1C and 6C to Top10-Rb bacteria.
- Starter for pBS1C3_MCS and for the positive colonies with the plasmid containing fragments 2C, 3C. 5C and 8C.
- Additional amplification for F4, F4* and F5.
- First Gibson assembly attempt. For more information – please read what Ilya wrote…
Lior
Noa
Evgeni
Shahar
Rachel
|


 "
"