Team:TU Munich/Project/Xanthohumol
From 2012.igem.org



Contents |
Xanthohumol
Background and principles
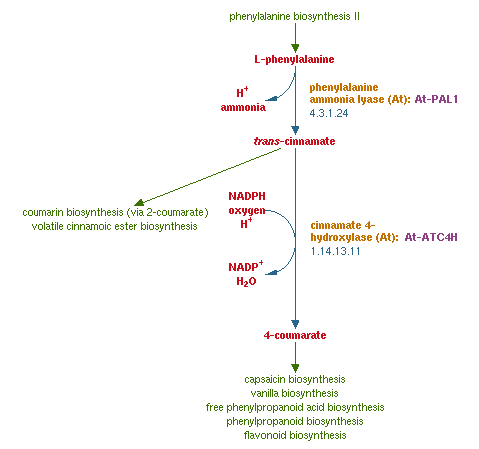
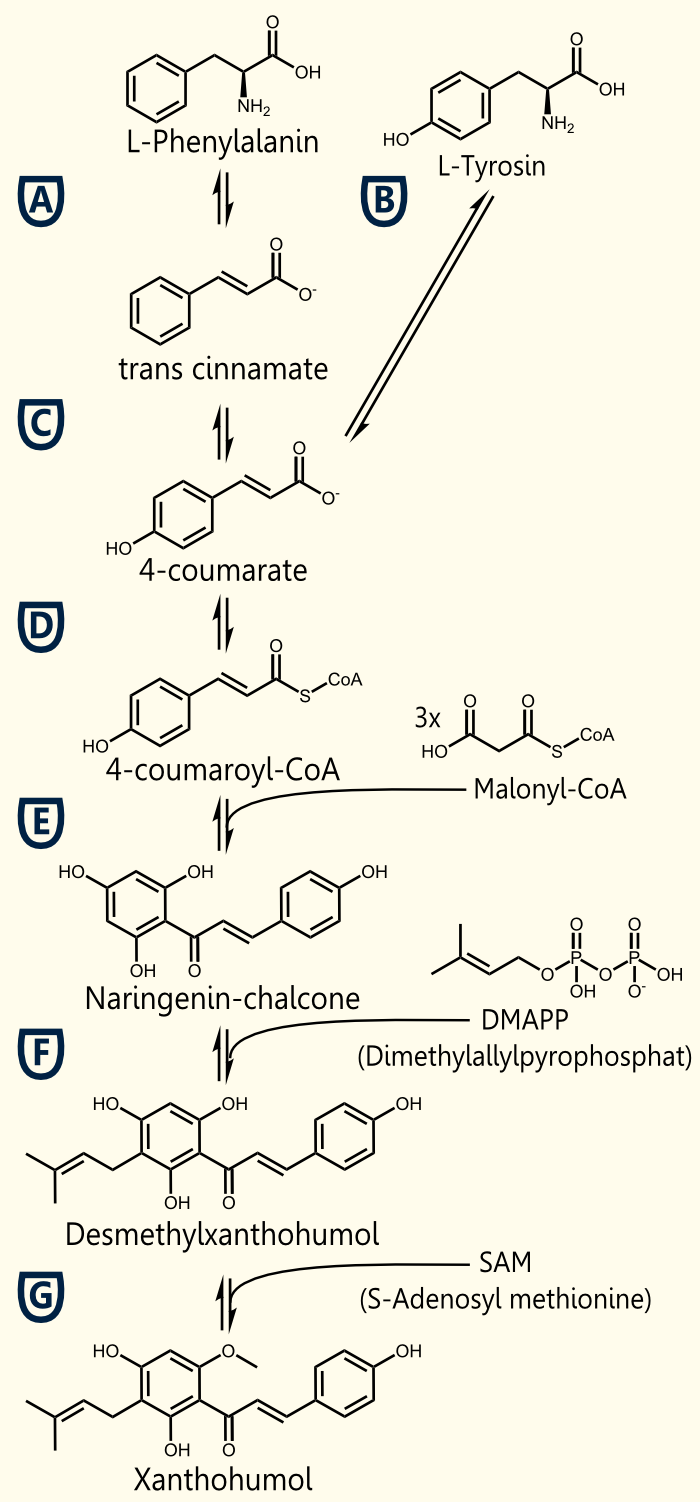
Plant secondary metabolites have proven or assumed beneficial properties and health promoting effects. Stilbenoids, flavonoids or lignins can result from 4-coumaroyl-coenzyme A, which is a nodal compound of phenylproponaoid metabolism in plants.
Biosynthesis:
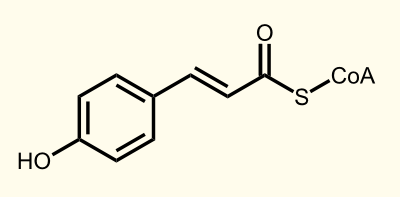
The biosynthetic pathway of 4-coumaroyl-coenzyme A starts with the conversion of L-Phenylalanine to cinnamate catalyzed by phenylalanin ammonia lyase (PAL). PAL also shows activity with converting tyrosine to p-coumarate, albeit to a lower efficiency. The cinnamate 4-hydroxylase (C4H) catalyzes the synthesis of p-hydroxycinnamate from cinnamate and 4-coumarate: CoA ligase (4CL) converts p-coumarate to its coenzyme-A ester, activating it for reaction with malonyl CoA Emmanouil Trantas et al., 2009.
The flavonoid biosynthetic pathway starts with the condensation of one molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA, yielding naringenin chalcone. This reaction is carried out by the enzyme chalcone synthase (CHS). Chalcone is isomerised to a flavanone by the enzyme chalcone flavanone isomerase (CHI). From these central intermediates, the pathway diverges into several side branches, each resulting in a different class of flavonoids, for example Xanthohumol.
Media:TUM12_Biosynthesis_of_Xanthohumol_(2).jpg
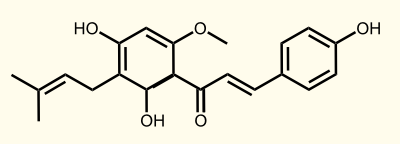
Our project will focus on the production of Xanthohumol, due to its characteristic as a cancer chemopreventive agent (see below).
Inhibition of the metabolic activation of procarcinogens:
2-amino-3-methylimidazo[4,5-f]quinolone, found in cooked meat, verified as a procarcinogen in an ames salmonella mutagenicity test. The inhibition is probably a result of an inhibition of the cytochrome P 450 enzymes Cyp1A1, Cyp1B1 and Cyp1A2 (phase 1 enzymes). But in order to achieve a clear inhibition, plasma concentrations of 1 µM would be necessary. In a study with male rats oral administration of xanthohumol (50 mg/kg) led to concentration maximums of 65 -180 nM after 4 h. Improved resorption of Xanthohumol could be a possible target for innovation (Yilmazer et al., 2001, Miranda et al., 2000, Henderson et al., 2000, Gerhauser et al., 2002).
Induction of carcinogen-detoxifying enzymes (phase 2 enzymes):
P450-activated carcinogens get conjugated to endogenous ligands (gluthathione, glucoronic acid, acetate and sulfate) by phase 2 enzymes to facilitate excretion. Therefore the induction of phase 2 enzymes should enhance the protection against carcinogenesis. Xanthohumol cat concentrations of 2.1-10.1 µM could induce quinone reductase (detoxification of quinones by ceonversion to hydroquinones which can be conjugated) in hepatoma Hepa 1c1c7 cells. It was shown that xanthohumol could selectively induce quinone reductase without causing a transcriptional activation of Cyp1A1. (Miranda et al., 2000, Gerhauser et al., 2002)
Inhibition of tumor growth at an early stage:
Xantohumol showed an inhibition of the proliferation of breast cancer (MCF-7) and ovarian cancer (A-2780) in vitro at IC50 values of 13 and 0.52 µM (Miranda et al., 1999). Furthermore xanthohumol can inhibit the endogenous prostaglandin synthesis through inhibition of cyclooxygenase (COX-1 and COX-2) with IC50 values of 17 and 42 µM. An increased prostaglandin production has been associated with the uncontrolled proliferation of tumor cells (Gerhauser et al., 2002). Pharmacokinetic studies for xanthohumol based on beverages with an xanthohumol content of 50 mg/l in humans are part of actual research activities. According to a scientist at the TA-XAN AG the first results will be published in June at a conference in Florenz.
Antioxidant activities:
Xanthohumol at 5 µM decreased conjugated diene formation as a measure for lipid peroxidation by more than 70 % after 5 h of incubation in an in vitro assay (protection of LDL from Cu2+ induced oxidation). Furthermore Xanthohumol was shown to scavenge hydroxyl-, peroxyl- and superoxide anion radicals (Miranda et al., 2000).
Idea
The idea is to perform a heterologous gene expression of all enzymes required for Xanthohumol biosynthesis in Saccharomyces cerevisiae. First, each enzyme should be expressed individually and the activities should be tested individually to ensure the functionality. Each gene should be inserted in a yeast expression vector under the control of a GAL10 promotor.
The final goal is the expression of all required genes in a single modified yeast to produce Xanthohumol out of the substrate L-Tyrosin.
General remarks
Proof of principle
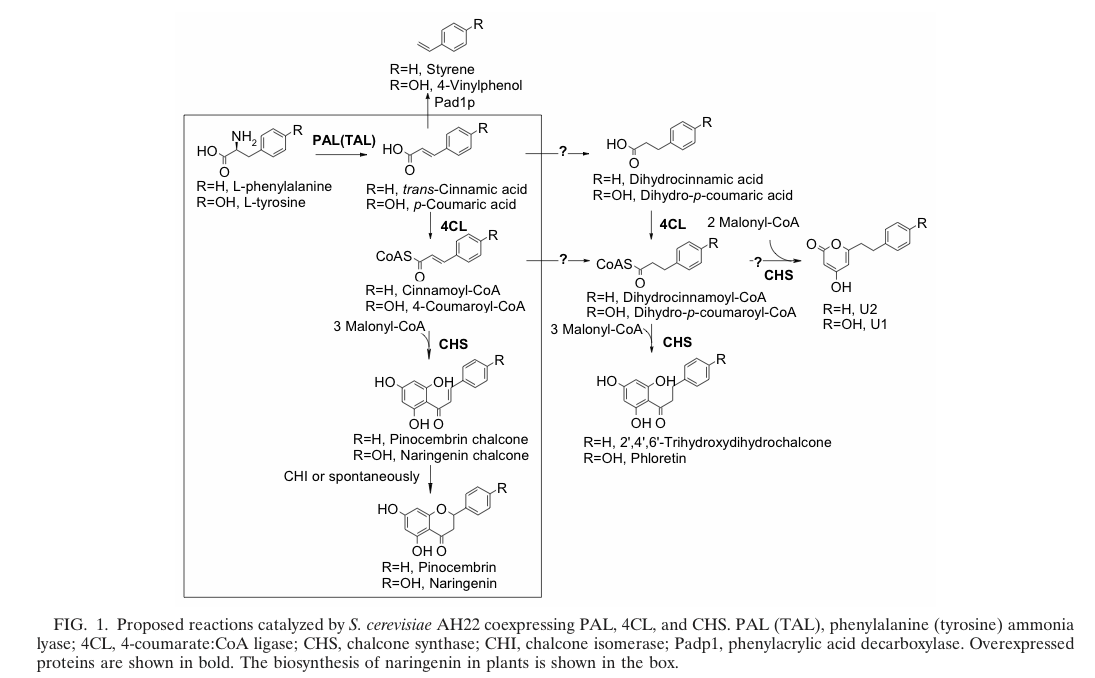
Jiang et al succeed in the biosynthesis of several flavonoids in Saccharomyces cerevisiae by the assembly of a plasmid which contains three required enzymes (pKS2µHyg-PAL-4CL-CHS). The activity of each enzyme was demonstrated and the presence of naringenin, which forms the product of the three enzymes(PAL, 4CL, CHS; see also picture on the right) was shown. Hanxiao Jiang et al., 2004
Necessary enzymes for the biosynthesis of xanthohumol
ENZYME 1: PAL = phenylalanine ammonia lyase: L-phenylalanin --> trans-cinnamate
ENZYME 2: 4CL = 4-coumarate - coenzym A ligase: 4-coumarate --> 4-coumaroyl-CoA
ENZYME 3: CHS = naringenin - chalcone synthase: 4-coumaroyl-CoA --> naringeninchalcone
ENZYME 4: APT = aromatic prenyltransferase: naringeninchalcone --> desmethylxanthohumol
ENZYME 5: OMT1 = chalcone O-methyltransferase: desmethylxanthohumol --> xanthohumol
Source: http://biocyc.org/META/NEW-IMAGE?type=NIL&object=PWY-5135
Enzymes - Sequences, Translation and Assays
Sequences of PAL, 4Cl and CHS come from Paper Hanxiao Jiang et al., 2004
Enzyme 1: phenylalanine ammonia lyase (PAL)
we only use the sequence from start- to stopcodon (bold): 1-2151
Sequence
>gi|18698155|emb|AX366866.1| Sequence 18 from Patent WO0208402
1 atggcaccct cgctcgactc gatctcgcac tcgttcgcaa acggcgtcgc atccgcaaag
61 caggctgtca atggcgcctc gaccaacctc gcagtcgcag gctcgcacct gcccacaacc
121 caggtcacgc aggtcgacat cgtcgagaag atgctcgccg cgccgaccga ctcgacgctc
181 gaactcgacg gctactcgct caacctcgga gacgtcgtct cggccgcgag gaagggcagg
241 cctgtccgcg tcaaggacag cgacgagatc cgctcaaaga ttgacaaatc ggtcgagttc
301 ttgcgctcgc aactctccat gagcgtctac ggcgtcacga ctggatttgg cggatccgca
361 gacacccgca ccgaggacgc catctcgctc cagaaggctc tcctcgagca ccagctctgc
421 ggtgttctcc cttcgtcgtt cgactcgttc cgcctcggcc gcggtctcga gaactcgctt
481 cccctcgagg ttgttcgcgg cgccatgaca atccgcgtca acagcttgac ccgcggccac
541 tcggctgtcc gcctcgtcgt cctcgaggcg ctcaccaact tcctcaacca cggcatcacc
601 cccatcgtcc ccctccgcgg caccatctct gcgtcgggcg acctctctcc tctctcctac
661 attgcagcgg ccatcagcgg tcacccggac agcaaggtgc acgtcgtcca cgagggcaag
721 gagaagatcc tgtacgcccg cgaggcgatg gcgctcttca acctcgagcc cgtcgtcctc
781 ggcccgaagg aaggtctcgg tctcgtcaac ggcaccgccg tctcagcatc gatggccacc
841 ctcgctctgc acgacgcaca catgctctcg ctcctctcgc agtcgctcac ggccatgacg
901 gtcgaagcga tggtcggcca cgccggctcg ttccacccct tccttcacga cgtcacgcgc
961 cctcacccga cgcagatcga agtcgcggga aacatccgca agctcctcga gggaagccgc
1021 tttgctgtcc accatgagga ggaggtcaag gtcaaggacg acgagggcat tctccgccag
1081 gaccgctacc ccttgcgcac gtctcctcag tggctcggcc cgctcgtcag cgacctcatt
1141 cacgcccacg ccgtcctcac catcgaggcc ggccagtcga cgaccgacaa ccctctcatc
1201 gacgtcgaga acaagacttc gcaccacggc ggcaatttcc aggctgccgc tgtggccaac
1261 accatggaga agactcgcct cgggctcgcc cagatcggca agctcaactt cacgcagctc
1321 accgagatgc tcaacgccgg catgaaccgc ggcctcccct cctgcctcgc ggccgaagac
1381 ccctcgctct cctaccactg caagggcctc gacatcgccg ctgcggcgta cacctcggag
1441 ttgggacacc tcgccaaccc tgtgacgacg catgtccagc cggctgagat ggcgaaccag
1501 gcggtcaact cgcttgcgct catctcggct cgtcgcacga ccgagtccaa cgacgtcctt
1561 tctctcctcc tcgccaccca cctctactgc gttctccaag ccatcgactt gcgcgcgatc
1621 gagttcgagt tcaagaagca gttcggccca gccatcgtct cgctcatcga ccagcacttt
1681 ggctccgcca tgaccggctc gaacctgcgc gacgagctcg tcgagaaggt gaacaagacg
1741 ctcgccaagc gcctcgagca gaccaactcg tacgacctcg tcccgcgctg gcacgacgcc
1801 ttctccttcg ccgccggcac cgtcgtcgag gtcctctcgt cgacgtcgct ctcgctcgcc
1861 gccgtcaacg cctggaaggt cgccgccgcc gagtcggcca tctcgctcac ccgccaagtc
1921 cgcgagacct tctggtccgc cgcgtcgacc tcgtcgcccg cgctctcgta cctctcgccg
1981 cgcactcaga tcctctacgc cttcgtccgc gaggagcttg gcgtcaaggc ccgccgcgga
2041 gacgtcttcc tcggcaagca agaggtgacg atcggctcga acgtctccaa gatctacgag
2101 gccatcaagt cgggcaggat caacaacgtc ctcctcaaga tgctcgctta gacactcttc
2161 ccactctcgc atcccttcca taccctatcc cgcctgcact cttaggactc gcttcttgtc
2221 ggactcggat ctcgcatcgc ttctttcgtt cttggctgcc tctctagacc gtgtccgtat
2281 tacctcgaga ttgtgaatac aagcagtacc catccacgca tccgataaat cagggagaga
2341 atctacgctt gcgggagctt cttgcgcata aactgtcgag tgcgggcgtt agtgcgaagt
2401 caacgaaggc gagtggcagc ggctcactac cgcctcgag
Translation
>gi|18698155|emb|AX366866.1| 1-2151
MAPSLDSISHSFANGVASAKQAVNGASTNLAVAGSHLPTTQVTQVDIVEKMLAAPTDSTLELDGYSLNLGDVVSAARKGRPVRVKDSDEIRSKIDKSVEFLRSQLSMSVYGVTTGFGGSADTRTEDAISLQKALLEHQLC GVLPSSFDSFRLGRGLENSLPLEVVRGAMTIRVNSLTRGHSAVRLVVLEALTNFLNHGITPIVPLRGTISASGDLSPLSYIAAAISGHPDSKVHVVHEGKEKILYAREAMALFNLEPVVLGPKEGLGLVNGTAVSASMAT LALHDAHMLSLLSQSLTAMTVEAMVGHAGSFHPFLHDVTRPHPTQIEVAGNIRKLLEGSRFAVHHEEEVKVKDDEGILRQDRYPLRTSPQWLGPLVSDLIHAHAVLTIEAGQSTTDNPLIDVENKTSHHGGNFQAAAVAN TMEKTRLGLAQIGKLNFTQLTEMLNAGMNRGLPSCLAAEDPSLSYHCKGLDIAAAAYTSELGHLANPVTTHVQPAEMANQAVNSLALISARRTTESNDVLSLLLATHLYCVLQAIDLRAIEFEFKKQFGPAIVSLIDQHF GSAMTGSNLRDELVEKVNKTLAKRLEQTNSYDLVPRWHDAFSFAAGTVVEVLSSTSLSLAAVNAWKVAAAESAISLTRQVRETFWSAASTSSPALSYLSPRTQILYAFVREELGVKARRGDVFLGKQEVTIGSNVSKIYE AIKSGRINNVLLKMLA
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Phenylalanine ammonia lyase | 2439bp bzw. 2151bp | ok | 6xNgoMIV (1438,1684,1852,1995,2329) | 0 AS<10% | [1] |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Phenylalanine ammonia lyase | XbaI, AgeI | Strep tag II | substrate: L-tyrosin, product: 4-coumarate |
Enzyme 2: 4-coumarate - coenzym A ligase (4CL)
we only use the sequence from start- to stopcodon (bold): 6-1691
>gi|609339|gb|U18675.1|ATU18675 Arabidopsis thaliana 4-coumarate--coenzyme A ligase (At4CL1) mRNA, complete cds
1 ttacaatggc gccacaagaa caagcagttt ctcaggtgat ggagaaacag agcaacaaca
61 acaacagtga cgtcattttc cgatcaaagt taccggatat ttacatcccg aaccacctat
121 ctctccacga ctacatcttc caaaacatct ccgaattcgc cactaagcct tgcctaatca
181 acggaccaac cggccacgtg tacacttact ccgacgtcca cgtcatctcc cgccaaatcg
241 ccgccaattt tcacaaactc ggcgttaacc aaaacgacgt cgtcatgctc ctcctcccaa
301 actgtcccga attcgtcctc tctttcctcg ccgcctcctt ccgcggcgca accgccaccg
361 ccgcaaaccc tttcttcact ccggcggaga tagctaaaca agccaaagcc tccaacacca
421 aactcataat caccgaagct cgttacgtcg acaaaatcaa accacttcaa aacgacgacg
481 gagtagtcat cgtctgcatc gacgacaacg aatccgtgcc aatccctgaa ggctgcctcc
541 gcttcaccga gttgactcag tcgacaaccg aggcatcaga agtcatcgac tcggtggaga
601 tttcaccgga cgacgtggtg gcactacctt actcctctgg cacgacggga ttaccaaaag
661 gagtgatgct gactcacaag ggactagtca cgagcgttgc tcagcaagtc gacggcgaga
721 acccgaatct ttatttccac agcgatgacg tcatactctg tgttttgccc atgtttcata
781 tctacgcttt gaactcgatc atgttgtgtg gtcttagagt tggtgcggcg attctgataa
841 tgccgaagtt tgagatcaat ctgctattgg agctgatcca gaggtgtaaa gtgacggtgg
901 ctccgatggt tccgccgatt gtgttggcca ttgcgaagtc ttcggagacg gagaagtatg
961 atttgagctc gataagagtg gtgaaatctg gtgctgctcc tcttggtaaa gaacttgaag
1021 atgccgttaa tgccaagttt cctaatgcca aactcggtca gggatacgga atgacggaag
1081 caggtccagt gctagcaatg tcgttaggtt ttgcaaagga accttttccg gttaagtcag
1141 gagcttgtgg tactgttgta agaaatgctg agatgaaaat agttgatcca gacaccggag
1201 attctctttc gaggaatcaa cccggtgaga tttgtattcg tggtcaccag atcatgaaag
1261 gttacctcaa caatccggca gctacagcag agaccattga taaagacggt tggcttcata
1321 ctggagatat tggattgatc gatgacgatg acgagctttt catcgttgat cgattgaaag
1381 aacttatcaa gtataaaggt tttcaggtag ctccggctga gctagaggct ttgctcatcg
1441 gtcatcctga cattactgat gttgctgttg tcgcaatgaa agaagaagca gctggtgaag
1501 ttcctgttgc atttgtggtg aaatcgaagg attcggagtt atcagaagat gatgtgaagc
1561 aattcgtgtc gaaacaggtt gtgttttaca agagaatcaa caaagtgttc ttcactgaat
1621 ccattcctaa agctccatca gggaagatat tgaggaaaga tctgagggca aaactagcaa
1681 atggattgtg atggatgatt tcaaccaaaa agcaaagatg atttcaatgt gtatatacat
1741 acaactgttt gacccaacca aggaaacaaa ctcatacgaa ccattgtctt ttgttgttgt
1801 tgttgttgtt gttgttgctg ttcttgcttg attcatgtaa tgagcctttg tgatgaaggt
1861 ggtttcttt
Translation
6-1691
MAPQEQAVSQVMEKQSNNNNSDVIFRSKLPDIYIPNHLSLHDYIFQNISEFATKPCLINGPTGHVYTYSDVHVISRQIAANFHKLGVNQNDVVMLLLPNCPEFVLSFLAASFRGATATAANPFFTPAEIAKQAKASNTKLIITEARYVDKIKPLQNDDGV VIVCIDDNESVPIPEGCLRFTELTQSTTEASEVIDSVEISPDDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYFHSDDVILCVLPMFHIYALNSIMLCGLRVGAAILIMPKFEINLLLELIQRCKVTVAPMVPPIVLAIAKSSETEKYDL SSIRVVKSGAAPLGKELEDAVNAKFPNAKLGQGYGMTEAGPVLAMSLGFAKEPFPVKSGACGTVVRNAEMKIVDPDTGDSLSRNQPGEICIRGHQIMKGYLNNPAATAETIDKDGWLHTGDIGLIDDDDELFIVDRLKELIKYKGFQVAPAELEALLIGH PDITDVAVVAMKEEAAGEVPVAFVVKSKDSELSEDDVKQFVSKQVVFYKRINKVFFTESIPKAPSGKILRKDLRAKLANGL
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Arabidopsis thaliana 4-coumarate--coenzyme A ligase | 1869 bp bzw. 1685 bp | 2xEcoRI (149-154, 309-314), 1x Spe1 (679-684) | ok after RFC10 | 0 AS<10% | [2] |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Arabidopsis thaliana 4-coumarate--coenzyme A ligase | XbaI, PstI | crude protein extraction | substrate: 4-coumarate, product: 4-coumaryl-CoA |
Enzyme 3: naringenin - chalcone synthase(CHS)
we only use the sequence from start- to stopcodon (bold): 76-1248
>gi|11096318|gb|AF315345.1| Hypericum androsaemum chalcone synthase mRNA, complete cds
1 aaactctgtc accacattat tgtaccttgt aacagcaagg tgcttaactg gttgatttaa
61 acataaacaa ggaagatggt gaccgtggaa gaagtcagga aggcgcagcg ggccgagggt
121 ccggccaccg tgatggccat cggaacggcc gtcccgccga actgcgttga ccaagcgacg
181 taccccgact attatttccg tatcaccaac agcgagcaca aggccgagct caaggagaag
241 ttccaacgca tgtgtgataa gtctcaaatc aagaaacgtt acatgtacct gaacgaggag
301 gtcctcaaag agaaccccaa tatgtgtgct tacatggcac cttctctgga tgctaggcaa
361 gacattgtgg tggtggaagt gcccaaacta ggtaaagagg cagcagttaa ggccatcaag
421 gaatggggcc agcctaagtc caagatcacc cacttggtct tttgcaccac tagtggagtg
481 gacatgcccg gggccgacta ccagctcacc aagctattgg gcctccgccc gtcggtgaag
541 cgcctcatga tgtaccagca gggctgcttt gccggtggca cggtcctccg tctcgccaag
601 gatctcgccg agaacaacaa gggtgcacgc gtccttgtcg tctgctcgga gatcacggcc
661 gttaccttcc gtgggcccac cgacactcac ctcgacagcc ttgtgggcca ggcattgttc
721 ggtgacggcg ctgccgccat catcatcggc tcggacccga tccccgaagt cgagaagccc
781 ttgttcgagc tggtctccgc agcccagacc attctaccgg acagtgaggg tgcgatagac
841 ggacatctcc gcgaggttgg gcttacattc cacttgctca aggatgttcc cgggttgatc
901 tctaagaacg ttgagaagag cctcactgag gccttcaaac cgttgggcat ttcagattgg
961 aactccctgt tttggatcgc ccacccaggc ggcccagcaa tcttggacca ggtagaggcc
1021 aagttgagcc tcaagcccga gaagctacgg gccacaaggc acgtactttc cgagtacgga
1081 aacatgtcta gtgcctgtgt gcttttcatc ttagacgaga tgaggaggaa gtccaaggaa
1141 gacgggctta agaccacagg ggaaggaatc gagtggggag tgctttttgg atttgggcct
1201 gggcttaccg ttgagaccgt tgtccttcac agtgtcgcca ttaactaggt caaggtcgtt
1261 gctttgcgtt ttttactttg ttgttgcctg taatattttc actacttggc gtcttttttt
1321 cactttctaa cttctaatgt tttacctctg ggtcaaacat atgtggtgca gtgaaaaact
1381 gaaaaaaaaa aaaaaaaaaa aa
Translation 76-1248
MVTVEEVRKAQRAEGPATVMAIGTAVPPNCVDQATYPDYYFRITNSEHKAELKEKFQRMCDKSQIKKRYMYLNEEVLKENPNMCAYMAPSLDARQDIVVVEVPKLGKEAAVKAIKEWGQPKSKITHLVFCTTSGVDMPGADYQLTKLLGLRPSVKRLMMY QQGCFAGGTVLRLAKDLAENNKGARVLVVCSEITAVTFRGPTDTHLDSLVGQALFGDGAAAIIIGSDPIPEVEKPLFELVSAAQTILPDSEGAIDGHLREVGLTFHLLKDVPGLISKNVEKSLTEAFKPLGISDWNSLFWIAHPGGPAILDQVEAKLSLK PEKLRATRHVLSEYGNMSSACVLFILDEMRRKSKEDGLKTTGEGIEWGVLFGFGPGLTVETVVLHSVAIN
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Hypericum androsaemum chalcone synthase | 1402 bp | 1xSpeI (469) | ok after RFC10 | 2 AS<10% |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Hypericum androsaemum chalcone synthase | XbaI, AgeI | Strep tag II | subtrate: |
Enzyme 4: aromatic prenyltransferase (APT)
we decided to have the gene synthesized
>gi|11096318|gb|(AB543053.1)| optimized sequence with restriction sites (XbaI and AgeI) (gene synthesis)
1 tctagatggctttgtcatccgtttcttcattttctttgggtaccaacccattcatctcca 61 tcccacataacaacaacaacttgaaggtttcttcctactgctgcaaatctaagtccagag 121 ttatcaactccactaactctaaacattgctccccaaacaacaacaacaacacttctaaca 181 agaccacccatttgttgggtttatacggtcaatcaagatgcttgttgaagccattgtctt 241 tcatctcttgcaacgatcaaagaggtaactctattagagcttccgcccaaattgaagata 301 gaccaccagaatctggtaacttgtctgctttgactaacgttaaggatttcgtttctgttt 361 gctgggaatacgttagaccatatactgctaagggtgttatcatttgctcctcttgtttgt 421 tcggtagagaattattggaaaacccaaacttgttctccagaccattgattttcagagcct 481 tgttgggtatgttggctattttgggttcttgtttttacaccgccggtatcaatcaaatct 541 tcgatatggatatcgacagaatcaacaagccagatttgccattggtttccggtagaattt 601 ctgttgaatctgcttggttgttgactttgtccccagctattattggtttcatcttgatct 661 tgaagttgaactccggtcctttgttgacctcattatactgtttggcaatcttgtccggta 721 ctatctattctgttccaccttttagatggaagaagaatccaattaccgccttcttgtgca 781 ttttgatgattcatgctggtttgaacttctccgtttactatgcttcaagagctgctttgg 841 gtttggcttttgcttggtcaccatctttttctttcattaccgctttcatcaccttcatga 901 ctttgactttggcttcctctaaggatttgtccgatattaacggtgatagaaagttcggtg 961 ttgaaactttcgctacaaaattgggtgctaagaacatcaccttgttaggtactggtttgt 1021 tattattgaactacgttgctgctatttccaccgctattatttggcctaaagctttcaagt 1081 ccaacatcatgttgttgtcccatgctatcttggccttttcattgatctttcaagctagag 1141 aattggacagaactaactacactccagaagcttgtaagtccttctacgaatttatctgga 1201 ttttgttctccgccgaatacgttgtttacttgttcatcaccggt
Translation
1-1236
MELSSVSSFSLGTNPFISIPHNNNNLKVSSYCCKSKSRVINSTNSKHCSPNNNNNTSNKTTHLLGLYGQSRCLLKPLSFISCNDQRGNSIRASAQIEDRPPESGNLSALTNVKDFVSVCWEYVRPYTAKGVIICSSCLFGRELLENPNLFSRPLIFRALL GMLAILGSCFYTAGINQIFDMDIDRINKPDLPLVSGRISVESAWLLTLSPAIIGFILILKLNSGPLLTSLYCLAILSGTIYSVPPFRWKKNPITAFLCILMIHAGLNFSVYYASRAALGLAFAWSPSFSFITAFITFMTLTLASSKDLSDINGDRKFGVE TFATKLGAKNITLLGTGLLLLNYVAAISTAIIWPKAFKSNIMLLSHAILAFSLIFQARELDRTNYTPEACKSFYEFIWILFSAEYVVYLFI
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Humulus lupulus aromatic prenyltransferase | 1236 bp | ok | ok | optimized by GeneArt |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Humulus lupulus aromatic prenyltransferase | XbaI, AgeI | Strep tag II | substrate: |
Enzyme 5: chalcone O-methyltransferase(OMT1)
we only use the sequence from start- to stopcodon (50-1108bold):
Humulus lupulus O-methyltransferase 1 (OMT1) mRNA, complete cds, Genbank EU309725.1
1 ggacacaatt caatctattt tacccaaaaa ataactaaga aagaccaata tggaatctct
61 aagaggccaa gaacagatat ggcaactcat gttcagcttt gtcgactcca tggccttgaa
121 atgcgccatc gagcttcgca ttgctgacat cattcactct catggcaaac ctataactct
181 ctcccaaata gcttctggca ttcgatcaaa ctccaactcc tccatatctc cgaatattcc
241 ttacctctct cgcatcatga gatttcttgt tcgaaagaat atcttcactg aacatcaaga
301 agataatgat gaggtgatct cattgtacgg gctaagtgat agctcgagat ggctgttgcg
361 ggattttaag tcaagcctgg ctcccatggt gctcatgcag actcatccat tgtcgatggc
421 ggtgtggcat ttccttgagg attatgtgag aaacagcagc aacactttcg aaaaggctca
481 cggttgtaac atttgggagt tttcctcagc caatccagat ttcaacaaga tcttcaacaa
541 tgccatggcg agtattgtgc caatatacat gggggctgtg ctttcaagtt ataaggatgg
601 tcttggttgt attaaaggaa cagtggtgga cgttgggggt ggtacgggcg gctccatatc
661 agagcttatg aaatattatc caaacatcaa agggattaac tttgacctgc cacatgtgat
721 tgccacagca ccggcattgg atggtgttac ccatattagt ggtgacatat tcgagtcaat
781 tcctagtgct gatgcggttt taatgaaggg tgtactacat tgcttcagcg atgaaaaatg
841 tgtaaaagta ttgagaaatt gtcgaaaagc aataacagac aaaaagaatg ggaagattat
901 cattttggag attgtgttgg acccaaccag caatcaaata tttgacgaga ctcgaatggt
961 gtacgattta ttgattccay tctttagtgg tggaaaagag agaactgagc ttgaatggaa
1021 aaggctatta aacgaggctg gttttacttc tatcaaaatc accaaaattc caattatacc
1081 tgctattatt gaggcctttc tagtgtgaca acrtcgatct atctatatat atataaacta
1141 ggttatgttg ctttcaacaa taagttccct atgtactgtt acggttatgt atggtttgct
1201 gtgattaata taatatgttg gc
Translation
50-1108
MESLRGQEQIWQLMFSFVDSMALKCAIELRIADIIHSHGKPITLSQIASGIRSNSNSSISPNIPYLSRIMRFLVRKNIFTEHQEDNDEVISLYGLSDSSRWLLRDFKSSLAPMVLMQTHPLSMAVWHFLEDYVRNSSNTFEKAHGCNIWEFSSANPDFNK IFNNAMASIVPIYMGAVLSSYKDGLGCIKGTVVDVGGGTGGSISELMKYYPNIKGINFDLPHVIATAPALDGVTHISGDIFESIPSADAVLMKGVLHCFSDEKCVKVLRNCRKAITDKKNGKIIILEIVLDPTSNQIFDETRMVYDLLIPXFSGGKERTE LEWKRLLNEAGFTSIKITKIPIIPAIIEAFLV
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Humulus lupulus O-methyltransferase 1 | 1058 | ok | ok | 2 AS<10% |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Humulus lupulus O-methyltransferase 1 | XbaI, AgeI | Strep tag II | subtrate: |
References
- C Gerhauser, A Alt, E Heiss, A Gamal-Eldeen, K Klimo, J Knauft, I Neumann, H.R Scherf, N Frank, H Bartsch, H Becker Cancer chemopreventive activity of xanthohumol, a natural product derived from hop Mol. Cancer Ther., 1 (2002), pp. 959–969
- M.C Henderson, C.L Miranda, J.F Stevens, M.L Deinzer, D.R Buhler In vitro inhibition of human P450 enzymes by prenylated flavonoids from hops, Humulus lupulus Xenobiotica, 30 (2000), pp. 235–251
- C.L Miranda, G.L Aponso, J.F Stevens, M.L Deinzer, D.R Buhler Prenylated chalcones and flavanones as inducers of quinone reductase in mouse Hepa 1c1c7 cells Cancer Lett., 149 (2000), pp. 21–29
- C.L Miranda, J.F Stevens, A Helmrich, M.C Henderson, R.J Rodriguez, Y.H Yang, M.L Deinzer, D.W Barnes, D.R Buhler Antiproliferative and cytotoxic effects of prenylated flavonoids from hops (Humulus lupulus) in human cancer cell lines Food Chem. Toxicol., 37 (1999), pp. 271–285
- C.L Miranda, J.F Stevens, V Ivanov, M McCall, B Frei, M.L Deinzer, D.R Buhler Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro J. Agric. Food Chem., 48 (2000), pp. 3876–3884
- C.L Miranda, Y.H Yang, M.C Henderson, J.F Stevens, G Santana-Rios, M.L Deinzer, D.R Buhler, Prenylflavonoids from hops inhibit the metabolic activation of the carcinogenic heterocyclic amine 2-amino-3-methylimidazo[4,5-f]quinoline, mediated by cDNA-expressed human CYP1A2 Drug Metab. Dispos., 28 (2000), pp. 1297–1302
- M Yilmazer, J.F Stevens, M.L Deinzer, D.R Buhler In vitro biotransformation of xanthohumol, a flavonoid from hops (Humulus lupulus), by rat liver microsomes Drug Metab. Dispos., 29 (2001), pp. 223–231
- M Yilmazer, J.F Stevens, D.R Buhler In vitro glucuronidation of xanthohumol, a flavonoid in hop and beer, by rat and human liver microsomes FEBS Lett., 491 (2001), pp. 252–256
 "
"