Team:Marburg SYNMIKRO
From 2012.igem.org
(Difference between revisions)
| (99 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Marburg_SYNMIKRO:Template:Header}} | {{:Team:Marburg_SYNMIKRO:Template:Header}} | ||
| - | < | + | |
| + | <html> | ||
| + | <div class="conbox60"> | ||
| + | </html> | ||
| + | == '''<html> </html>Recombinator:''' <br/> <html><sup> </sup></html>An intelligent Genetically Engineered Slot Machine (iGESM) == | ||
<html> | <html> | ||
| - | <div | + | <div style="text-align:justify; padding-left:2px; padding-right:2px;"> |
| - | + | The vertebrate immune system produces billions of different antibodies. This diversity is generated by random VDJ-recombination of a limited number of antibody subfragments. This inspired us to construct an automatic recombination system in <i>E. coli</i> that generates large numbers of novel proteins by combinatorial fusion of functional domains. We used the site-specific DNA recombinase Gin of bacteriophage Mu that depends on the presence of a DNA enhancer element for efficient recombination. This allowed us to construct a system, called “The Recombinator”, which automatically shuts down after successful recombination. We visualized the randomizing function of our genetically engineered slot machine by combining fluorescent colors with cellular localization domains. By scaling up the number of recombination modules and functional domains our system will be able to generate a multitude of new proteins. We envision that “The Recombinator” will serve as a tool to create novel enzymatic activities for innovative drug design, environmental detoxification and metabolic engineering. | |
| - | This is a | + | |
</div> | </div> | ||
| - | |||
| - | |||
</div> | </div> | ||
| - | <div | + | <div style="background-color:white;" class="conbox28"> |
| - | + | <h3 style="z-index:1000;" >Team mixer</h3><a style="float:right; position: relative; top:-25px; z-index:1000;" href="https://2012.igem.org/Team:Marburg_SYNMIKRO/TeamProfile">[Team members]</a> | |
| + | <style type="text/css"> | ||
| + | #people{ | ||
| + | border: hidden 0px; | ||
| + | border-bottom: black dotted 1px; | ||
| + | background: transparent; | ||
| + | color: darkblue; | ||
| + | width:139px; | ||
| + | text-align: right; | ||
| + | |||
| + | } | ||
| + | |||
| + | #footer-box { | ||
| + | position: relative; | ||
| + | top:-10px; | ||
| + | z-index: 1000; | ||
| + | } | ||
| + | |||
| + | #container{ | ||
| + | height: 578px; | ||
| + | width: 262px; | ||
| + | position: relative; | ||
| + | /* margin-left:50%;*/ | ||
| + | /* left:-284px;*/ | ||
| + | } | ||
| + | |||
| + | |||
| + | |||
| + | #image{ | ||
| + | height: 578px; | ||
| + | width: 262px; | ||
| + | float:left; | ||
| + | position:relative; | ||
| + | |||
| + | } | ||
| + | |||
| + | |||
| + | #hd{ | ||
| + | height:91px; | ||
| + | width: 163px; | ||
| + | } | ||
| + | |||
| + | #td{ | ||
| + | position: relative; | ||
| + | height:197px; | ||
| + | } | ||
| + | |||
| + | #ld{ | ||
| + | position: relative; | ||
| + | height:235px; | ||
| + | } | ||
| + | |||
| + | .btn { | ||
| + | background:#a00; | ||
| + | color:#fff !important; | ||
| + | border: 1px solid #900; | ||
| + | padding: 4px 8px; | ||
| + | border-radius: 4px; | ||
| + | -moz-border-radius: 4px; | ||
| + | -webkit-border-radius: 4px; | ||
| + | margin: 20px; | ||
| + | text-transform: uppercase; | ||
| + | text-decoration: none !important; | ||
| + | font: bold 16px Verdana, sans-serif; | ||
| + | text-shadow: 0 0 1px #000; | ||
| + | box-shadow: 0 2px 2px #aaa; | ||
| + | -moz-box-shadow: 0 2px 2px #aaa; | ||
| + | -webkit-box-shadow: 0 2px 2px #aaa; | ||
| + | cursor:pointer; | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | <script src="http://malsup.github.com/jquery.cycle.all.js"></script> | ||
| + | <script type="text/javascript"> | ||
| + | var intran; | ||
| + | intran = 3500; | ||
| + | rantim = setInterval("random();", intran); | ||
| + | var timer; | ||
| + | jQuery(function($) { | ||
| + | timer = setTimeout(blnk, 0); | ||
| + | }); | ||
| + | |||
| + | |||
| + | function blnk() { | ||
| + | $(".blink").css({opacity: 0}). | ||
| + | animate({opacity: 1}, 300, "linear"). | ||
| + | animate({opacity: 0}, 300, "linear", | ||
| + | function() { | ||
| + | timer = setTimeout(blnk, 0); | ||
| + | }); | ||
| + | } | ||
| + | |||
| + | |||
| + | infos = new Array( | ||
| + | |||
| + | "<b>Daniel Kleinsorge</b><br><br>text", | ||
| + | "<b>Oliver Schauer</b><br><br>text", | ||
| + | "<b>Prof. Michael Bölker</b><br><br>text", | ||
| + | "<b>Marian Vogt</b><br><br>text", | ||
| + | "<b>Agnieszka Zarzycka</b><br><br>text", | ||
| + | "<b>Oliver Leicht</b><br><br>text", | ||
| + | "<b>Margarita Derbenev</b><br><br>text", | ||
| + | "<b>Elisabeth Ignatz</b><br><br>text", | ||
| + | "<b>Julian Pechstein</b><br><br>text", | ||
| + | "<b>Johannes Schuh</b><br><br>text", | ||
| + | "<b>Max Diesner</b><br><br>text", | ||
| + | "<b>Svetlana Uscherenko</b><br><br>text" | ||
| + | ); | ||
| + | |||
| + | var numpeople=14; | ||
| + | var headnum=0; | ||
| + | var torsonum=0; | ||
| + | var legsnum=0; | ||
| + | var i=0; | ||
| + | var headimages=""; | ||
| + | var torsoimages=""; | ||
| + | var legsimages=""; | ||
| + | |||
| + | $(function() { | ||
| + | for (i=0; i<numpeople; i++) | ||
| + | { | ||
| + | headimages=headimages + "<img border='0' src='http://web383.mis60.de/inprogress/teambilder/" + i + "a.png' width='163' height='91'> "; | ||
| + | torsoimages=torsoimages + "<img border='0' src='http://web383.mis60.de/inprogress/teambilder/" + i + "b.png' width='163' height='197'> "; | ||
| + | legsimages=legsimages + "<img border='0' src='http://web383.mis60.de/inprogress/teambilder/" + i + "c.png' width='163' height='235'> "; | ||
| + | } | ||
| + | document.getElementById("s1").innerHTML=(headimages); | ||
| + | document.getElementById("s2").innerHTML=(torsoimages); | ||
| + | document.getElementById("s3").innerHTML=(legsimages); | ||
| + | |||
| + | |||
| + | $('#s1').cycle({ | ||
| + | fx: 'scrollHorz', | ||
| + | speed: 300, | ||
| + | timeout: 0, | ||
| + | }); | ||
| + | $('#s2').cycle({ | ||
| + | fx: 'scrollHorz', | ||
| + | speed: 300, | ||
| + | timeout: 0, | ||
| + | }); | ||
| + | $('#s3').cycle({ | ||
| + | fx: 'scrollHorz', | ||
| + | speed: 300, | ||
| + | timeout: 0, | ||
| + | }); | ||
| + | |||
| + | |||
| + | }); | ||
| + | |||
| + | function headleft() | ||
| + | { | ||
| + | if(headnum==0){ | ||
| + | headnum=(numpeople-1); | ||
| + | $('#s1').cycle('prev'); | ||
| + | } | ||
| + | else{ | ||
| + | headnum=(headnum-1); | ||
| + | $('#s1').cycle(headnum); | ||
| + | } | ||
| + | document.getElementById("infobox").innerHTML=("Ist das ein Bug oder ein Feature?"); | ||
| + | } | ||
| + | |||
| + | function headright() | ||
| + | { | ||
| + | if(headnum==(numpeople-1)){ | ||
| + | headnum=0; | ||
| + | $('#s1').cycle('next'); | ||
| + | } | ||
| + | else{ | ||
| + | headnum=(headnum+1); | ||
| + | $('#s1').cycle(headnum); | ||
| + | } | ||
| + | document.getElementById("infobox").innerHTML=("Wird allse live berechnet!"); | ||
| + | } | ||
| + | |||
| + | function torsoleft() | ||
| + | { | ||
| + | if(torsonum==0){ | ||
| + | torsonum=(numpeople-1); | ||
| + | $('#s2').cycle('prev'); | ||
| + | } | ||
| + | else{ | ||
| + | torsonum=(torsonum-1); | ||
| + | $('#s2').cycle(torsonum); | ||
| + | } | ||
| + | document.getElementById("infobox").innerHTML=("Das stand doch eben noch nicht hier??"); | ||
| + | } | ||
| + | |||
| + | function torsoright() | ||
| + | { | ||
| + | if(torsonum==(numpeople-1)){ | ||
| + | torsonum=0; | ||
| + | $('#s2').cycle('next'); | ||
| + | } | ||
| + | else{ | ||
| + | torsonum=(torsonum+1); | ||
| + | $('#s2').cycle(torsonum); | ||
| + | } | ||
| + | document.getElementById("infobox").innerHTML=("Hmm ja was schreibt man jetzt dazu?"); | ||
| + | } | ||
| + | |||
| + | function legsleft() | ||
| + | { | ||
| + | if(legsnum==0){ | ||
| + | legsnum=(numpeople-1); | ||
| + | $('#s3').cycle('prev'); | ||
| + | } | ||
| + | else{ | ||
| + | legsnum=(legsnum-1); | ||
| + | $('#s3').cycle(legsnum); | ||
| + | } | ||
| + | document.getElementById("infobox").innerHTML=("Hmm ja was schreibt man jetzt dazu?"); | ||
| + | } | ||
| + | |||
| + | function legsright() | ||
| + | { | ||
| + | if(legsnum==(numpeople-1)){ | ||
| + | legsnum=0; | ||
| + | $('#s3').cycle('next'); | ||
| + | } | ||
| + | else{ | ||
| + | legsnum=(legsnum+1); | ||
| + | $('#s3').cycle(legsnum); | ||
| + | } | ||
| + | document.getElementById("infobox").innerHTML=("Machts gut und danke für den Fisch!"); | ||
| + | } | ||
| + | |||
| + | |||
| + | function random() | ||
| + | { | ||
| + | |||
| + | clearInterval(rantim) | ||
| + | rantim = setInterval("random();", intran); | ||
| + | |||
| + | headnum=Math.round(Math.random()*(numpeople-1)); | ||
| + | $('#s1').cycle(headnum); | ||
| + | |||
| + | torsonum=Math.round(Math.random()*(numpeople-1)); | ||
| + | $('#s2').cycle(torsonum); | ||
| + | |||
| + | legsnum=Math.round(Math.random()*(numpeople-1)); | ||
| + | $('#s3').cycle(legsnum); | ||
| + | |||
| + | document.getElementById("infobox").innerHTML=("Hmm ja was schreibt man jetzt dazu?"); | ||
| + | |||
| + | } | ||
| + | |||
| + | function setall(num) | ||
| + | { | ||
| + | $('#s1').cycle(num);$('#s2').cycle(num);$('#s3').cycle(num); | ||
| + | headnum=num; | ||
| + | torsonum=num; | ||
| + | legsnum=num; | ||
| + | |||
| + | document.getElementById("infobox").innerHTML=infos[num]; | ||
| + | } | ||
| + | |||
| + | window.onload = random; | ||
| + | </script> | ||
| + | |||
| + | |||
| + | |||
| + | <div id="container"> | ||
| + | |||
| + | <div id="image"> | ||
| + | <center> | ||
| + | <div id="hd"> | ||
| + | <div id="s1" class="pics"> | ||
</div> | </div> | ||
| + | |||
</div> | </div> | ||
| - | |||
| - | < | + | <div id="td"> |
| + | <div id="s2" class="pics"> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | <div id="ld"> | ||
| + | <div id="s3" class="pics"> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | <div> | ||
| + | <br> | ||
| + | <a onclick="random(); " class="btn"><span class="blink">Recombine!</span></a> | ||
| + | </center> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| - | + | <html> | |
| - | + | <div style="top:345px; position: absolute; width:61.5%; height:299px;" class="conbox60"> | |
| - | + | </html> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
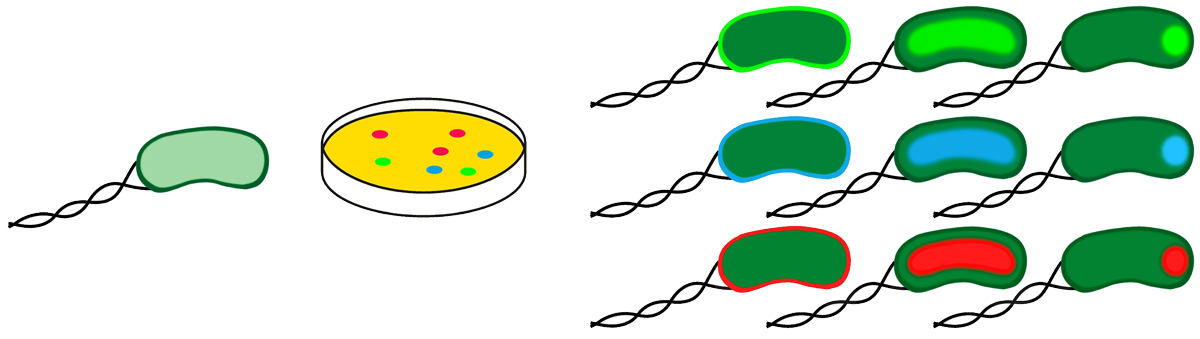
| - | + | == Bacterial combinatorics == | |
| - | + | As proof-of-principle of our recombinator system we created combinatorial fusions of bacterial intracellular localization domains with different fluorescent proteins. We expected to generate a mixture of bacteria that glow in different colors and at different positions in all possible combinations. | |
| - | + | <br/> | |
| - | + | <br/> | |
| - | + | [[File:Syn_MR_bunte_Maus.png|center|420px]] | |
| - | + | ||
| - | + | Here, we have dealt only with few modules, but with additional recombination modules, the number of possible combinations raises exponentially. | |
| - | + | <html> | |
| - | + | </div> | |
| - | + | <div class="conboxb"> | |
| - | + | </html> | |
| - | + | [[File:Syn_MR_Logo.png|link=http://www.uni-marburg.de|left|400px]][[File:Syn_MR_synmikro_farbe.gif|link=http://www.synmikro.com/en/|right|400px]] | |
| + | <html> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 13:25, 23 September 2013

Recombinator:
An intelligent Genetically Engineered Slot Machine (iGESM)
The vertebrate immune system produces billions of different antibodies. This diversity is generated by random VDJ-recombination of a limited number of antibody subfragments. This inspired us to construct an automatic recombination system in E. coli that generates large numbers of novel proteins by combinatorial fusion of functional domains. We used the site-specific DNA recombinase Gin of bacteriophage Mu that depends on the presence of a DNA enhancer element for efficient recombination. This allowed us to construct a system, called “The Recombinator”, which automatically shuts down after successful recombination. We visualized the randomizing function of our genetically engineered slot machine by combining fluorescent colors with cellular localization domains. By scaling up the number of recombination modules and functional domains our system will be able to generate a multitude of new proteins. We envision that “The Recombinator” will serve as a tool to create novel enzymatic activities for innovative drug design, environmental detoxification and metabolic engineering.
Bacterial combinatorics
As proof-of-principle of our recombinator system we created combinatorial fusions of bacterial intracellular localization domains with different fluorescent proteins. We expected to generate a mixture of bacteria that glow in different colors and at different positions in all possible combinations.
Here, we have dealt only with few modules, but with additional recombination modules, the number of possible combinations raises exponentially.
 "
"