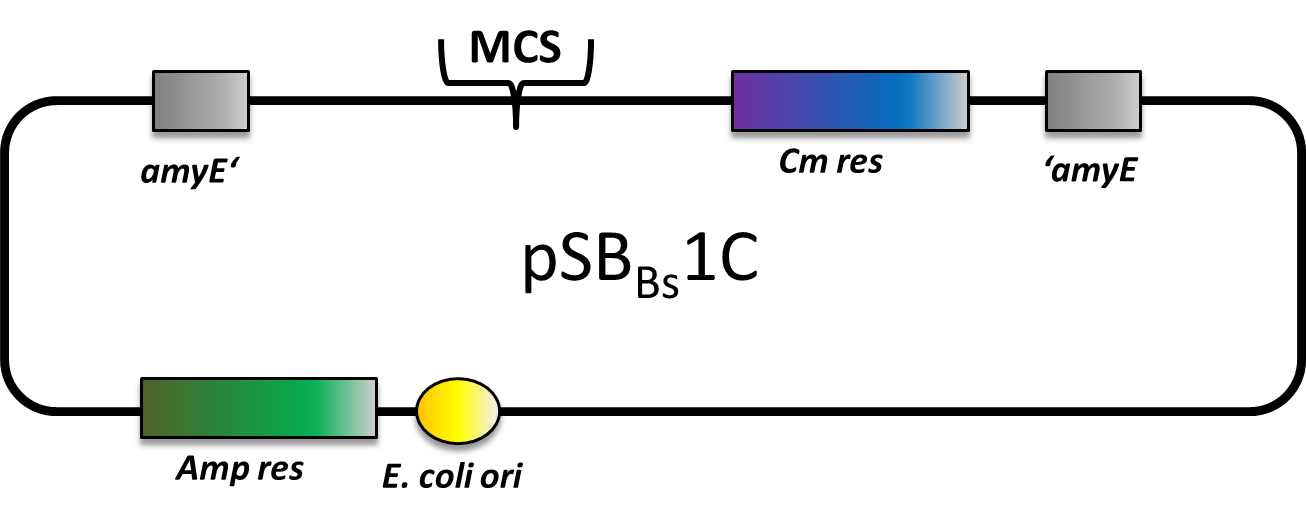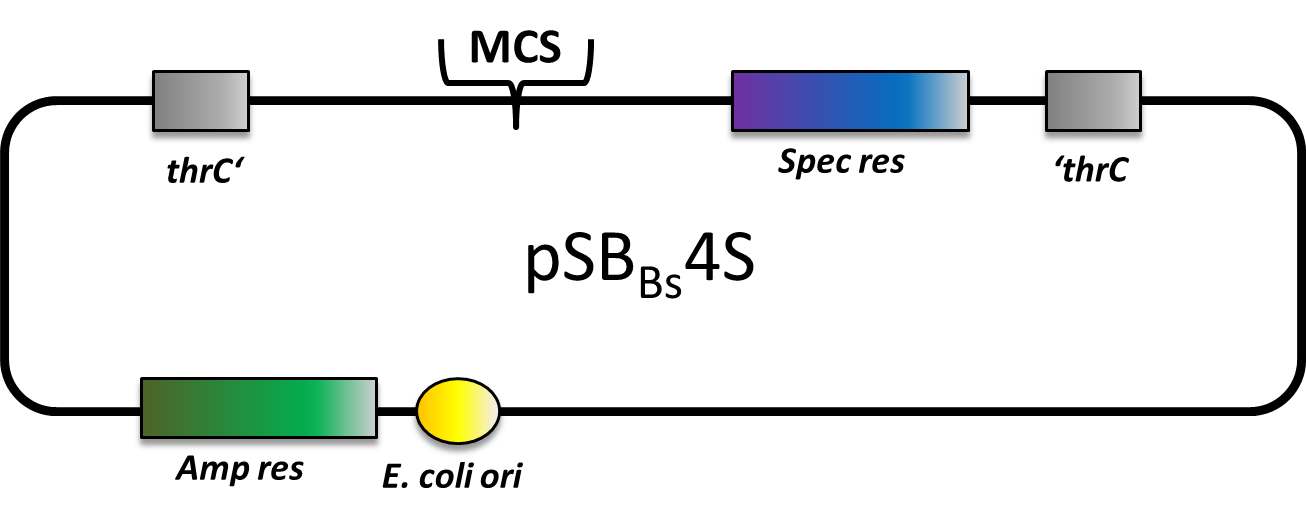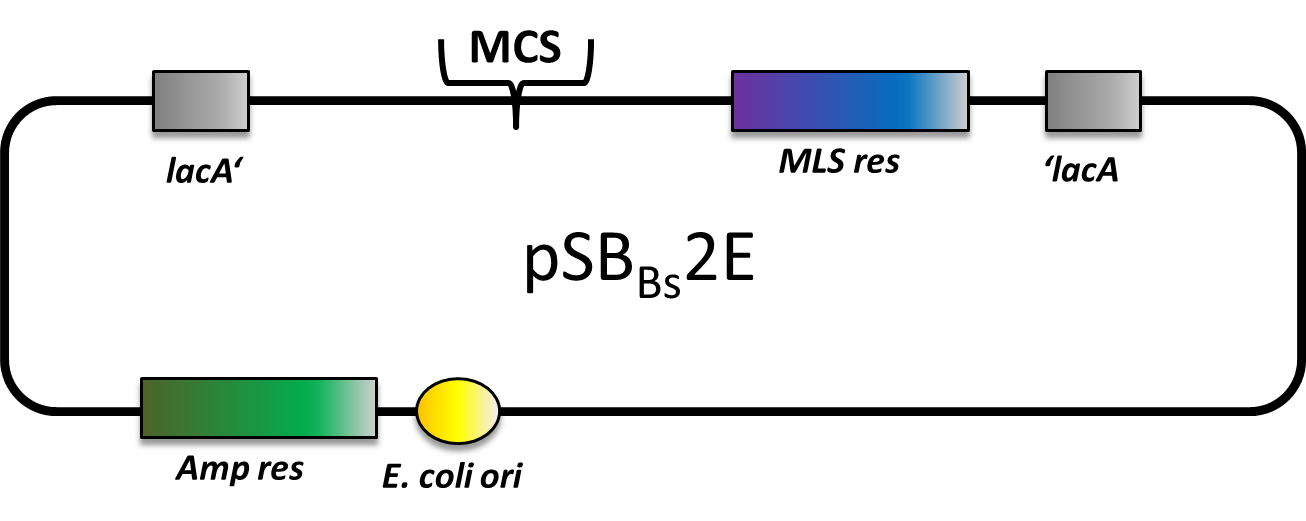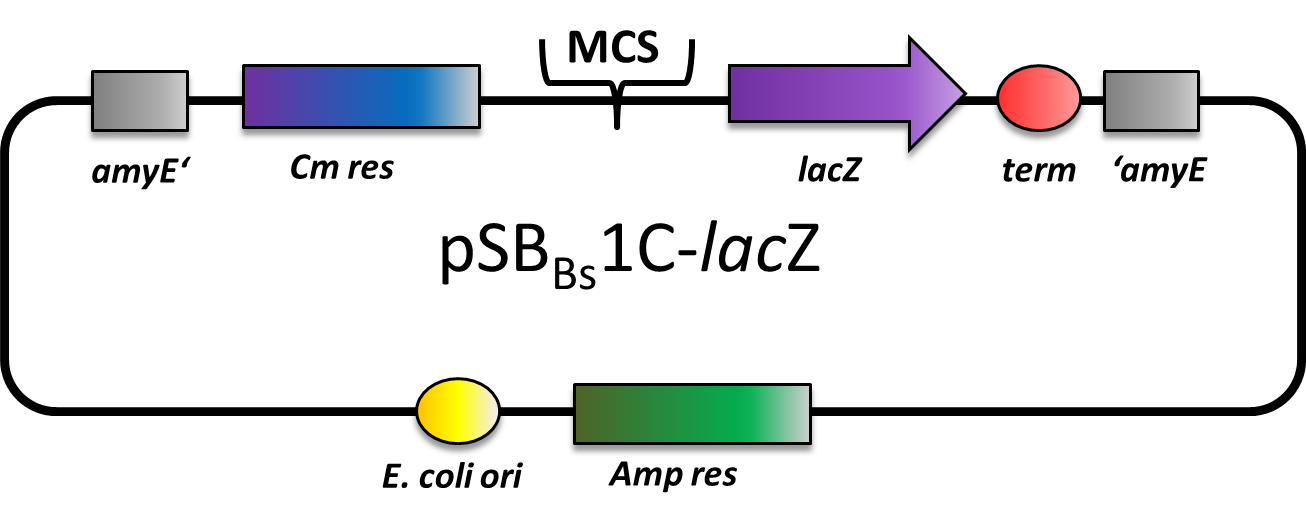Team:LMU-Munich/Data/Vectors
From 2012.igem.org
| Line 90: | Line 90: | ||
<div class="box"> | <div class="box"> | ||
===pSB<sub>''Bs''</sub>1C=== | ===pSB<sub>''Bs''</sub>1C=== | ||
| - | |||
[[File:LMU-Munich-PSBBs1C.png|400px|left]] | [[File:LMU-Munich-PSBBs1C.png|400px|left]] | ||
| - | pSB<sub>Bs</sub>1C is one of our empty vectors. It integrates into the ''amy''E locus of ''Bacillus subtilis'' and carries a chloramphenicol resistance as well as an ampicillin resistance for ''E. coli''. In between the two recombination sites, there is the constitutively expressed Cm resistance and the multiple cloning site which contains RFP to simplify the insertion of your BioBrick. | + | <p align="justify"> |
| + | pSB<sub>Bs</sub>1C is one of our empty vectors. It integrates into the ''amy''E locus of ''Bacillus subtilis'' and carries a chloramphenicol resistance as well as an ampicillin resistance for ''E. coli''. In between the two recombination sites, there is the constitutively expressed Cm resistance and the multiple cloning site which contains RFP to simplify the insertion of your BioBrick.</p> | ||
[[Image:LMU Firstspore.jpg|280px|link=Team:LMU-Munich/Spore_Coat_Proteins|left]][[File:LMU-Munich-starchplate.JPG | 300px|right]] | [[Image:LMU Firstspore.jpg|280px|link=Team:LMU-Munich/Spore_Coat_Proteins|left]][[File:LMU-Munich-starchplate.JPG | 300px|right]] | ||
| + | <p align="justify"> | ||
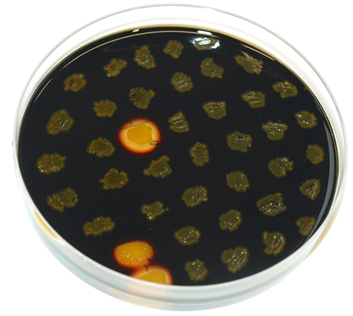
The plasmid is then transformed into ''B. subtilis'' and checked via the resistance for "integration at all" and via a starch test for "integration into the right locus". Correct integration results in loss of halo due to disruption of the amylase gene. | The plasmid is then transformed into ''B. subtilis'' and checked via the resistance for "integration at all" and via a starch test for "integration into the right locus". Correct integration results in loss of halo due to disruption of the amylase gene. | ||
| - | + | </p> | |
| + | <p align="justify"> | ||
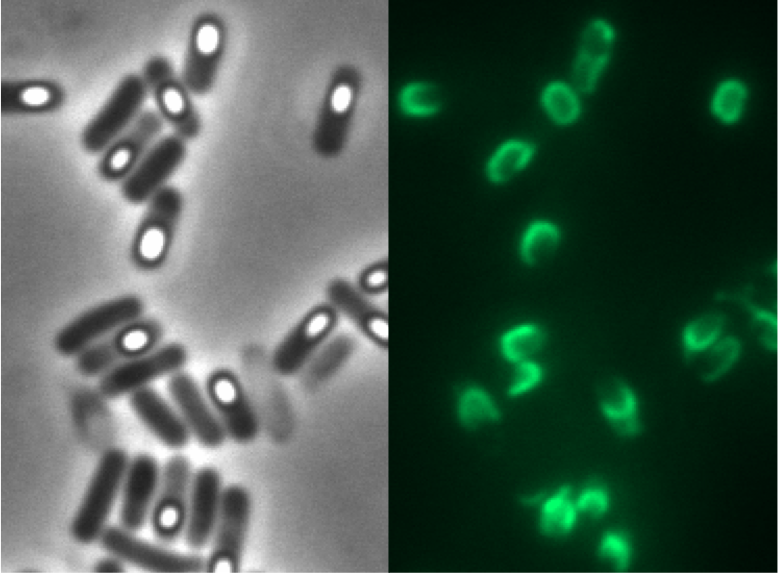
We tested this vector with various inserts, for example the final construct of our Sporobeads with GFP. The starch test demonstrated that the plasmid inserted correctly in the ''amy''E locus and fluorescence microscopy revealed the functionality of the construct. | We tested this vector with various inserts, for example the final construct of our Sporobeads with GFP. The starch test demonstrated that the plasmid inserted correctly in the ''amy''E locus and fluorescence microscopy revealed the functionality of the construct. | ||
| Line 104: | Line 106: | ||
<div class="box"> | <div class="box"> | ||
===pSB<sub>''Bs''</sub>4S=== | ===pSB<sub>''Bs''</sub>4S=== | ||
| - | + | ||
[[File:LMU-Munich-PSBBs4S.png|400px|left]] | [[File:LMU-Munich-PSBBs4S.png|400px|left]] | ||
[[File:LMU-Munich-Thrplate.jpg | 300px|left]] | [[File:LMU-Munich-Thrplate.jpg | 300px|left]] | ||
| - | pSB<sub>Bs</sub>4S is also an empty vector with only the multiple cloning site and the spectinomycin resistance gene in between the homologous regions. This vector integrates into the ''thr''C locus of ''B. subtilis''. | + | <p align="justify"> |
| - | + | pSB<sub>Bs</sub>4S is also an empty vector with only the multiple cloning site and the spectinomycin resistance gene in between the homologous regions. This vector integrates into the ''thr''C locus of ''B. subtilis''. </p> | |
| + | <p align="justify"> | ||
We tested the integration of this plasmid with the threonine test and a construct from the '''Suicide'''switch as an insert. | We tested the integration of this plasmid with the threonine test and a construct from the '''Suicide'''switch as an insert. | ||
<br> | <br> | ||
| Line 119: | Line 122: | ||
<div class="box"> | <div class="box"> | ||
===pSB<sub>''Bs''</sub>2E=== | ===pSB<sub>''Bs''</sub>2E=== | ||
| - | |||
[[File:LMU-Munich-PSBBs2E.png|400px|left]] | [[File:LMU-Munich-PSBBs2E.png|400px|left]] | ||
| + | <p align="justify"> | ||
pSB<sub>Bs</sub>2E is our third empty vector, aiming to provide a set of three empty vectors with three different integration loci and three different compatible resistance casseettes for full flexibility in their combination. | pSB<sub>Bs</sub>2E is our third empty vector, aiming to provide a set of three empty vectors with three different integration loci and three different compatible resistance casseettes for full flexibility in their combination. | ||
| - | + | </p> | |
| + | <p align="justify"> | ||
This vector is still a work in progress, but currently only the multiple cloning site is missing. Of course, it is not tested yet. | This vector is still a work in progress, but currently only the multiple cloning site is missing. Of course, it is not tested yet. | ||
| Line 130: | Line 134: | ||
<div class="box"> | <div class="box"> | ||
===pSB<sub>''Bs''</sub>1C-<i>lac</i>Z=== | ===pSB<sub>''Bs''</sub>1C-<i>lac</i>Z=== | ||
| - | |||
[[File:LMU-Munich-PSBBs1C-lacZ.png|400px|left]] | [[File:LMU-Munich-PSBBs1C-lacZ.png|400px|left]] | ||
[[File:Englisch_Auswertung_PliaI.png|right|300px]] | [[File:Englisch_Auswertung_PliaI.png|right|300px]] | ||
| - | pSB<sub>Bs</sub>1C-<i>lac</i>Z is a reporter vector, which contains the multiple cloning site upstream of a functional reporter device, consisting of ribosome binding site, β-galactosidase gene ''lac''Z and terminator. Also, a chloramphenicol resistance is in between the two flanking homolgy regions. Upon chloramphenicol selection, this vector integrates into the ''amy''E locus, which can be verified by the starch test. | + | <p align="justify"> pSB<sub>Bs</sub>1C-<i>lac</i>Z is a reporter vector, which contains the multiple cloning site upstream of a functional reporter device, consisting of ribosome binding site, β-galactosidase gene ''lac''Z and terminator. Also, a chloramphenicol resistance is in between the two flanking homolgy regions. Upon chloramphenicol selection, this vector integrates into the ''amy''E locus, which can be verified by the starch test.</p> |
| - | + | <p align="justify"> | |
This vector was used for several [2012.igem.org/Team:LMU-Munich/Data#Anderson_Promoter_Evaluation promoter evaluations]. | This vector was used for several [2012.igem.org/Team:LMU-Munich/Data#Anderson_Promoter_Evaluation promoter evaluations]. | ||
</p> | </p> | ||
| Line 156: | Line 159: | ||
<div class="box"> | <div class="box"> | ||
===pSB<sub>''Bs''</sub>4S-P<sub><i>Xyl</i></sub>=== | ===pSB<sub>''Bs''</sub>4S-P<sub><i>Xyl</i></sub>=== | ||
| - | <p align="justify"> <font size="5" color=" | + | <p align="justify"> <font size="5" color="FF0000">CAUTION: DOES NOT WORK! CANNOT BE TRANSFORMED INTO ''B. SUBTILIS''</font> |
[[File:LMU-Munich-PSBBs4S-Pxyl.png|400px|left]]</p> | [[File:LMU-Munich-PSBBs4S-Pxyl.png|400px|left]]</p> | ||
| Line 171: | Line 174: | ||
<div class="box"> | <div class="box"> | ||
===pSB<sub>''Bs''</sub>0K-P<sub><i>spac</i></sub>=== | ===pSB<sub>''Bs''</sub>0K-P<sub><i>spac</i></sub>=== | ||
| - | <font size="5" color=" | + | <font size="5" color="FF0000">CAUTION: THIS VECTOR DOES NOT WORK AS EXPECTED! THE PROMOTER IS STRONG CONSTITUTIVE INSTEAD OF IPTG-INDUCIBLE!</font> |
[[File:LMU-Munich-PSBBs0K-Pspac.png|400px|left]] | [[File:LMU-Munich-PSBBs0K-Pspac.png|400px|left]] | ||
<p align="justify"> | <p align="justify"> | ||
| Line 216: | Line 219: | ||
</p> | </p> | ||
<p align="justify"> | <p align="justify"> | ||
| - | Possible reason for these results are: a dysfunctional Lac-repressor, or point mutations in the operator. This needs to be checked by sequencing. | + | Possible reason for these results are: a dysfunctional Lac-repressor, or point mutations in the operator. This needs to be checked by sequencing.</p> |
</div> | </div> | ||
| Line 236: | Line 239: | ||
</p> | </p> | ||
| - | |||
</div> | </div> | ||
Revision as of 20:32, 26 September 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Bacillus subtilis vectors
For the detailed protocols, please see our protocol section.
Overview
| Name BioBrick | E. coli res. | B. subt. res. | Insertion locus | Description | Cloning status | Works? |
|---|---|---|---|---|---|---|
| pSBBs1C (BBa_K823023) | Amp | Cm | amyE | empty | ||
| pSBBs4S (BBa_K823022) | Amp | Spec | thrC | empty | ||
| pSBBs2E (BBa_K823027) | Amp | MLS | lacA | empty | in progress | not tested |
| pSBBs1C-lacZ (BBa_K823021) | Amp | Cm | amyE | lacZ reporter | ||
| pSBBs3C-luxABCDE (BBa_K823025) | Amp | Cm | sacA | luxABCDE reporter | ||
| pSBBs4S-PXyl (BBa_K823024) | Amp | Spec | thrC | Xylose-promoter | refuses transformation | |
| pSBBs0K-Pspac (BBa_K823026) | Amp | Kan | replicative | IPTG-promoter | unexpected | |
| Sporovector (BBa_K823054) | Amp | Spec | thrC | to make Sporobeads | not tested |
pSBBs1C
pSBBs1C is one of our empty vectors. It integrates into the amyE locus of Bacillus subtilis and carries a chloramphenicol resistance as well as an ampicillin resistance for E. coli. In between the two recombination sites, there is the constitutively expressed Cm resistance and the multiple cloning site which contains RFP to simplify the insertion of your BioBrick.
The plasmid is then transformed into B. subtilis and checked via the resistance for "integration at all" and via a starch test for "integration into the right locus". Correct integration results in loss of halo due to disruption of the amylase gene.
We tested this vector with various inserts, for example the final construct of our Sporobeads with GFP. The starch test demonstrated that the plasmid inserted correctly in the amyE locus and fluorescence microscopy revealed the functionality of the construct.
pSBBs4S
pSBBs4S is also an empty vector with only the multiple cloning site and the spectinomycin resistance gene in between the homologous regions. This vector integrates into the thrC locus of B. subtilis.
We tested the integration of this plasmid with the threonine test and a construct from the Suicideswitch as an insert.
pSBBs2E
pSBBs2E is our third empty vector, aiming to provide a set of three empty vectors with three different integration loci and three different compatible resistance casseettes for full flexibility in their combination.
This vector is still a work in progress, but currently only the multiple cloning site is missing. Of course, it is not tested yet.
pSBBs1C-lacZ
pSBBs1C-lacZ is a reporter vector, which contains the multiple cloning site upstream of a functional reporter device, consisting of ribosome binding site, β-galactosidase gene lacZ and terminator. Also, a chloramphenicol resistance is in between the two flanking homolgy regions. Upon chloramphenicol selection, this vector integrates into the amyE locus, which can be verified by the starch test.
This vector was used for several [2012.igem.org/Team:LMU-Munich/Data#Anderson_Promoter_Evaluation promoter evaluations].
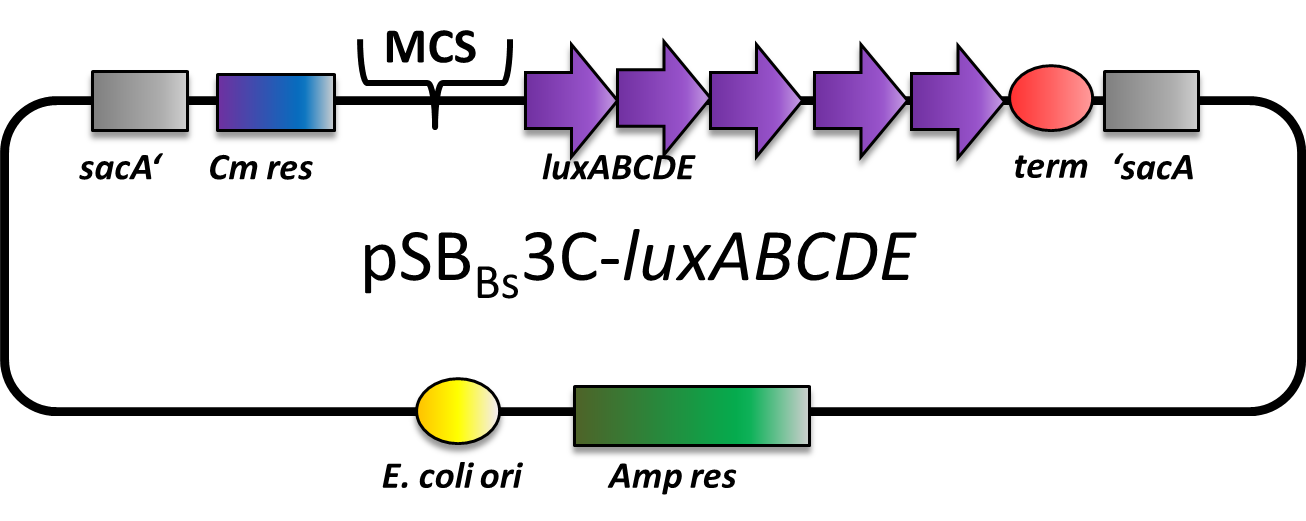
pSBBs3C-luxABCDE
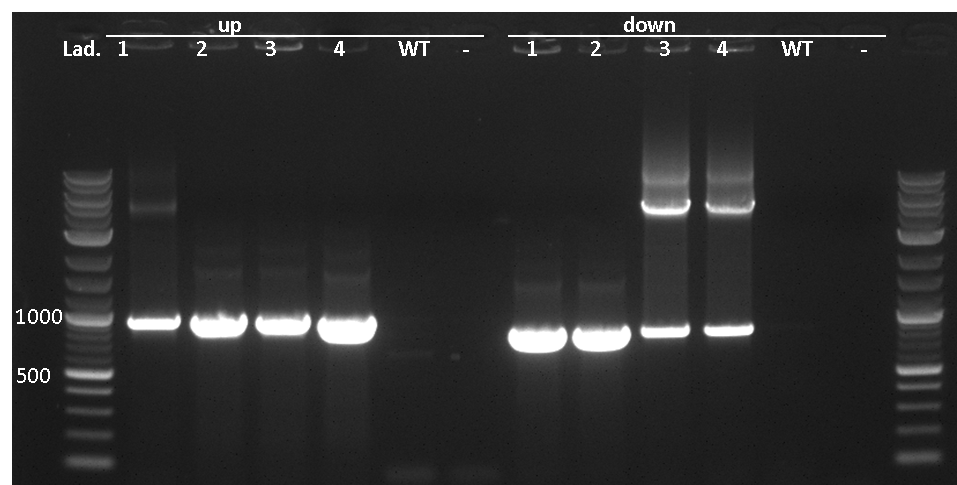
pSBBs3C-luxABCDE is our second reporter vector. It contains the multiple cloning site upstream of a functional reporter device, consisting of the lux-operon and a downstream terminator. Upon chloramphenicol selection, this vector integrates, this vector integrates into the sacA locus of B. subtilis by double homologous recombination.
Correct integration is verified by colony PCR. One primer is located outside the integration site and its respective partner is located on the inegrated part of the vector, facing outwards. gDNA from W168 as a negative control should not give a PCR product.
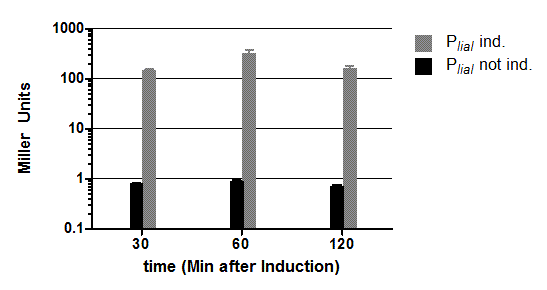
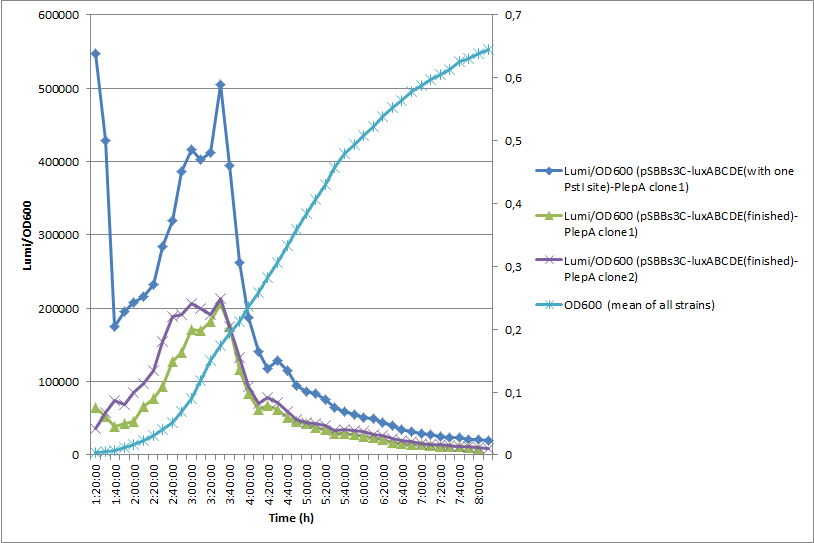
A pre-version of this vector with one forbidden PstI site was used for most promoter evaluations (All graphs that you can find in the Data section of the promoters derive from this pre-version). To proof that our final version also works, we did another experiment where we compared the luminescence of PlepA in the pre- and final version of pSBBs3C-luxABCDE. Here you can see the results of this experiment:
In this experiment, the finished vector showed about half of the activity throughout the measurement, hence while the absolute luminescence values are lower, the overall dynamics are unaffected.
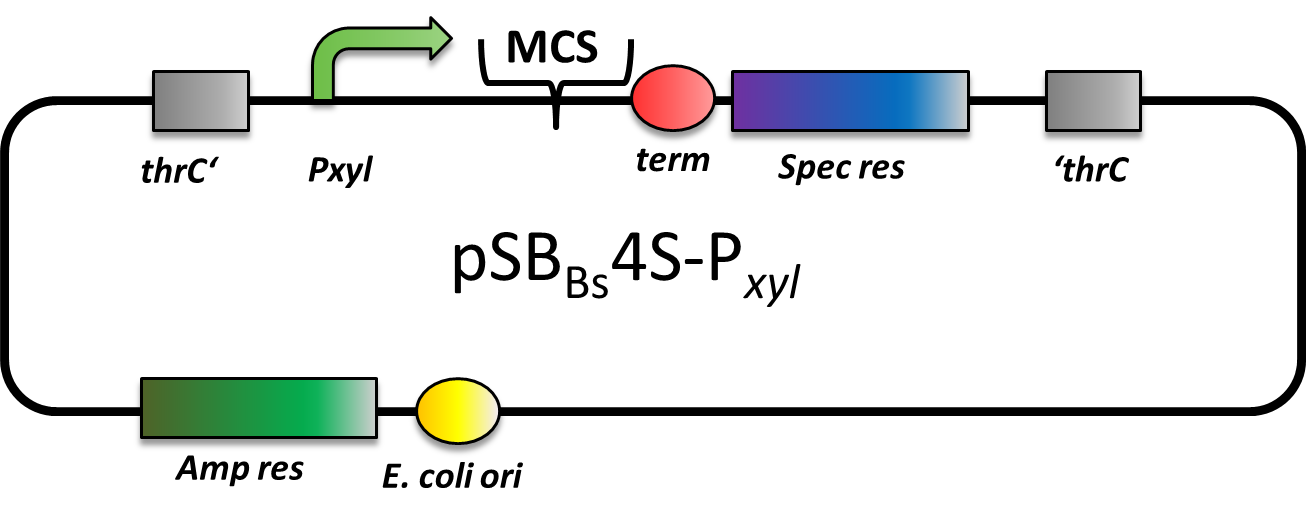
pSBBs4S-PXyl
CAUTION: DOES NOT WORK! CANNOT BE TRANSFORMED INTO B. SUBTILIS
pSBBs4S-PXyl is an integrative expression vector. Upstream of the multiple cloning site, there is a xylose-inducible promoter, Pxyl, followed by a terminator downstream. In the absence of xylose, Pxyl is inhibited by the repressor XylR, which is expressed in B. subtilis and not part of this vector. For selection, a spectinomycin resistance cassette is located in between the recombination sites. This vector integrates into the thrC locus, which can be checked by the threonine test.
However, we could not manage to transform this vector into B. subtilis, although we tried three times. Moreover, the team from Groningen also tested this vector, but again was not able to transform it into B. subtilis, either.
There are two possible reasons for the failure: First, a defective resistance cassette or second, the presence of a forbidden restriction site preventing correct linearization prior to transformation.
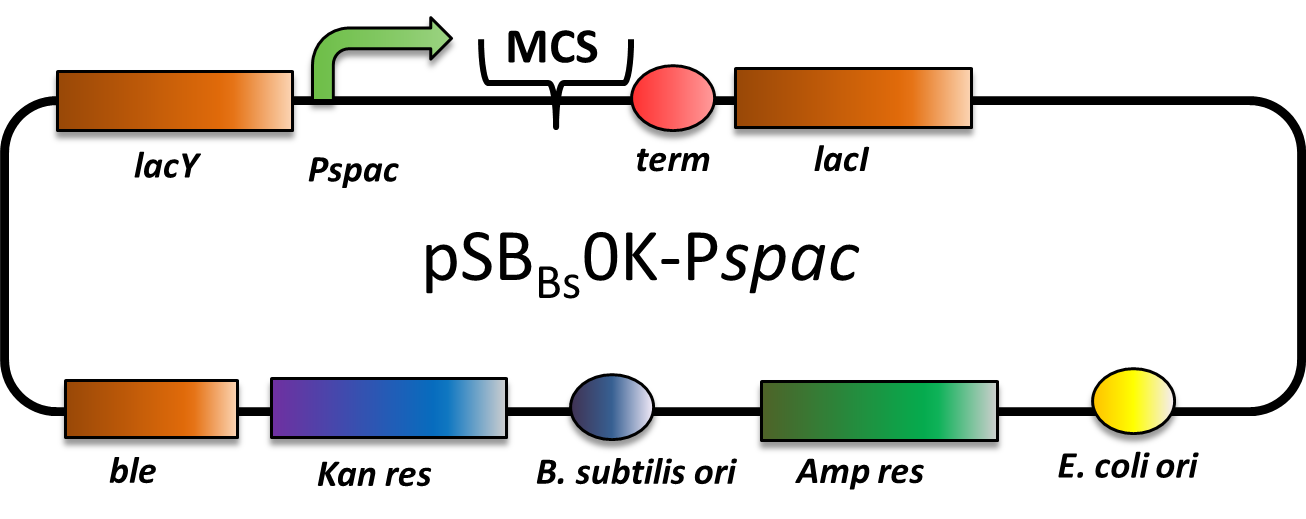
pSBBs0K-Pspac
CAUTION: THIS VECTOR DOES NOT WORK AS EXPECTED! THE PROMOTER IS STRONG CONSTITUTIVE INSTEAD OF IPTG-INDUCIBLE!
pSBBs0K-Pspac is a replicative expression vector with a kanamycin resistance. The IPTG -inducible promoter Pspac is followed by the multiple cloning site and a terminator. Moreover, it contains lacY, encoding aβ-Galactosid-Permease, and lacI, encoding the Lac-repressor. In the presence of IPTG, LacI is released from the promoter and the gene of interest is expressed.
The ble gene encodes the bleomycin resisitance protein (BRP), which can be used for by bleomycin selection in E. coli and B. subtilis. We did not use this resistance.
The presence of the vector can be checked by colony PCR with the primers: CTACATCCAGAACAACCTCTGC and TTCGGAAGGAAATGATGACCTC. We did not perfom the colony PCR because we selected for functionality of our lacZ-construct (see figure below) on plates with X-Gal and IPTG, and got blue colonies.
|
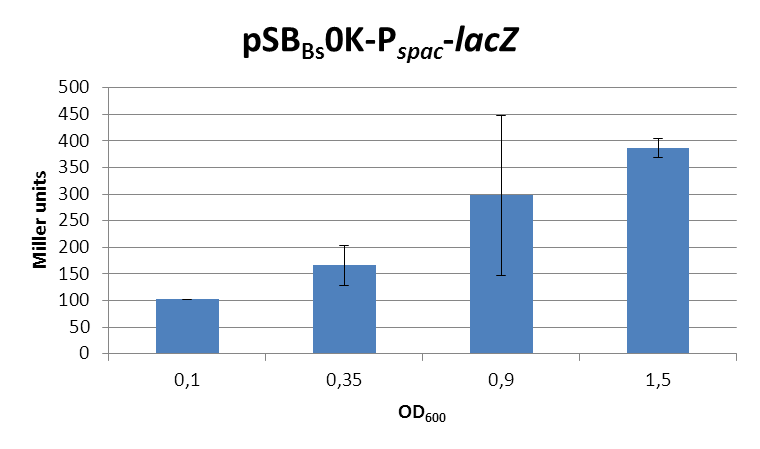
This expression vector was tested by insertion of lacZ into the multiple cloning site, transformation into W168 and ONPG assays. Overnight cultures were diluted 1:100 in LB-medium and incubated at 37°C, 230 rpm. Cells were harvested in 2 ml reaction tubes and frozen. For the induction assay, a culture of about OD600=0.9 was split into 3 ml aliquots in test tubes with different IPTG-concentrations and incubated for another hour. (Same results were obtained for OD600=0.3, data not shown.)
|
In summary, the figures show that the expression is uniformly very strong [in comparison to the native B. subtilis promoters], even without addition of IPTG. It does change depending on the growth phase, with a maximum strength in the stationary phase.
This vector was designed for the inducible overexpression of proteins, but is constitutively active.
Possible reason for these results are: a dysfunctional Lac-repressor, or point mutations in the operator. This needs to be checked by sequencing.
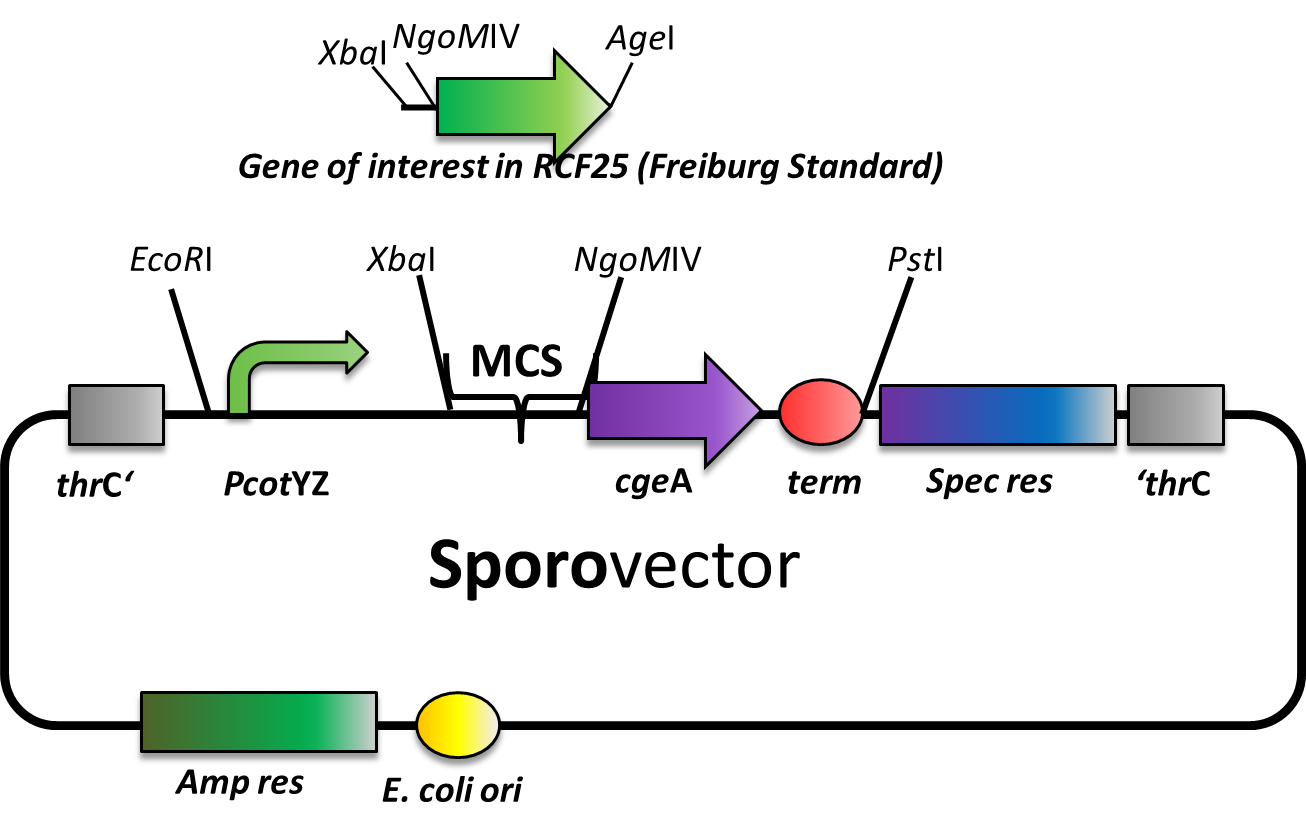
Sporovector
Our Sporovector is specially designed to give you the opportunity to easily create Sporobeads with any fusion protein you can think of. The part between the EcoRI and PstI is available as BioBrick in pSB1C3 and can be cloned in any of our empty B. subtilis vectors. We also offer it “precloned” in pSBBs4S, which in this version lacks the NgoMIV restriction sites.
The Sporovector was created by ligation of three cut PCR-products with special overhangs (PcotYZ, mRFP and cgeA-B0014). It is designed to allow N-terminal fusions of genes in Freiburg standard (RFC25). Towards that end, the gene to be inserted must be cut with XbaI and AgeI and the vector must be digested with XbaI and NgoMIV. This removes the RFP cassette and allows easy selection for the integration of the new insert. After ligation, there is a scar of six nucleotides between both genes but no restriction site left.
Expression of the gene fusion is controlled by the PcotYZ-promoter, which was the strongest in our promoter evaluations. The C-terminal part of the resulting fusion gene encodes CgeA, one of the two known spore crust proteins.
The vector is finished but has not yet been tested with an insert. We are working on three additional Sporovectors to also offer C and N-terminal fusions with both cotZ and cgeA.
 "
"