Alternative selectable and counter-selectable markers:
Levansucrase (sacB)
Background
SacB is the levansucrase enzyme from Bacillus subtilis (Gay, Coq, Strinmetz, Ferrari, & Hoch, 1983) which converts sucrose into fructose polymers which are lethal to Esherichia coli (French & Kowal, 2010). This part was deposited into the Registry by Team Edinburgh 2010 and can be used as a counter selectable marker (French & Kowal, 2010). Our aim is to improve the part by assessing its counter selection efficiency.
Cloning
The plac-RFP fragment was obtained from the standard BioBrick plasmid and inserted in front of the sacB BioBrick. Method. The construct was confirmed with sequencing.
PCR of pSB1K3 plasmid (Kanamycin resistance) with psBNX3 insF2 forward primer (specific for BioBrick prefix) and dsred r2 reverse primer (specific for RFP) was prepared in order to obtain the plac-RFP fragment.
Figure 1: DNA gel of the PCR product from pSB1K3 amplification with primers specific to the BioBrick prefix and RFP. The band is abound 1 kb which corresponds to the expected size of plac-RFP.
Close the plasmid.
The plac-RFP PCR product was purified and digested with EcoRI HF and SpeI. The sacB BioBrick deposited in 2010 (French & Kowal, 2010) was digested with EcoRI and XbaI. These were ligated together after purification. E.coli cells were transformed with the ligation. The red transformants were minipreped, digested with EcoRI HF in order to linearise them and with EcoRI HF and PstI in order to check the size of the insert.
Figure 2: DNA gel of miniprepped red clones of linearized plac-RFP-SacB ligation transformants. The band is around 4.5 kb which corresponds to pSB1C3 (2kb)+ +
Figure 3: The same clones were digested with EcoRI HF and PstI to check the size of the insert. The band is around 2.5 kb which corresponds to SacB (1.5 kb)+ plac-rfp (1kb).
Close the method.
Sequencing results
Forward primer:
Ctttaaaaaaaatcccttagctttcgctaaggtgatttctggaattcgcggccgcttctagagcaatac
gcaaaccgtttcaccccgcgcgttggccgattcattaatgcagctggcacgacaggtttcccgactgga
aagcgggcagtgagcgcaacgcaattaatgtgagttagctcactcattaggcaccccaggctttacact
ttatgcttccggctcgtatgttgtgtggaattgtgagcggataacaatttcacacatactagataaaga
ggagaaatactagatggcttcctccgaagacgttatcaaagagttcatgcgtttcaaagttcttatgga
aggttccgttaactgtcactagttcgaaatcgaaggtgaatgtgaaggtcgtccgtactaaggtaccca
gactgctaaactgaaagttactaaag
Reverse primer:
aggggccttaaacataaacttttcggttttagaaaagggcagggtggtgacaccttgcccttttttgcc
ggactgcagctactagtaatttatttgttaactgttaattgtccttgttcaaggatgctgtctttgaca
acagatgttttcttgcctttgatgttcagcaggaagcttggcgcaaacgttgattgtttgtctgcgtaa
aatcctctgtttgtcatatagcttgtaatcacgacattgtttcctttcgcttgaggtacagcgaagtgt
gagtaattaaaggttacatcgttaggatcaagatccatttttaacacatggcctgttttgttcagcggc
ttgtatgggccatttaaagaattagaaactttaccaagcatgttaatatcgttagacttatttccgtca
atccttatttttgatccgcgggagtcatttaacaggtaccatttgccgttcattttattttcgttcgcg
cgtctatttctttttgttactttgttttatgcaatcacgttttcattccttttttaattttgtatcatcgt
Close the sequencing results.
The idea of placing sacB under the control of the lac promoter is to create kanamycin-independent control. Moreover, by changing the IPTG levels, the level of selection can be controlled.
One of the weaknesses of sacB selection often mentioned in the literature is that sacB loss-of-function mutants also survive, and this may occur at higher frequency than loss of the cassette. RFP is added to ensure that cells which have lost the cassette (should be white because of loss of RFP) can be distinguished from cells which have lost SacB function (should be red as RFP is still present). This would allow us to assess the counter-selection efficiency of SacB.
We also prepared a Kan-plac-RFP-sacB selection-counterselection cassette.
Characterisation
The growth of SacB (bottom) and a control (top) were tested by adding 1.3 g solid sucrose plus 0.5 ml sterile water into a well in the middle of the plate, as shown in Figure 4 below..
Figure 4: A quick test of SacB transformants' growth in the presence of sucrose. The growth of SacB (bottom) is inhibited near the well in comparison to the control (top).
Citrobacter freundii
SacB and control (pSB1C3 only) transformants were streaked onto LB plates (with either cml40 or cml15) and solid sucrose was added to the regions indicated by the crosses or circles in Figure 5. The plates were incubated at 37°C overnight.
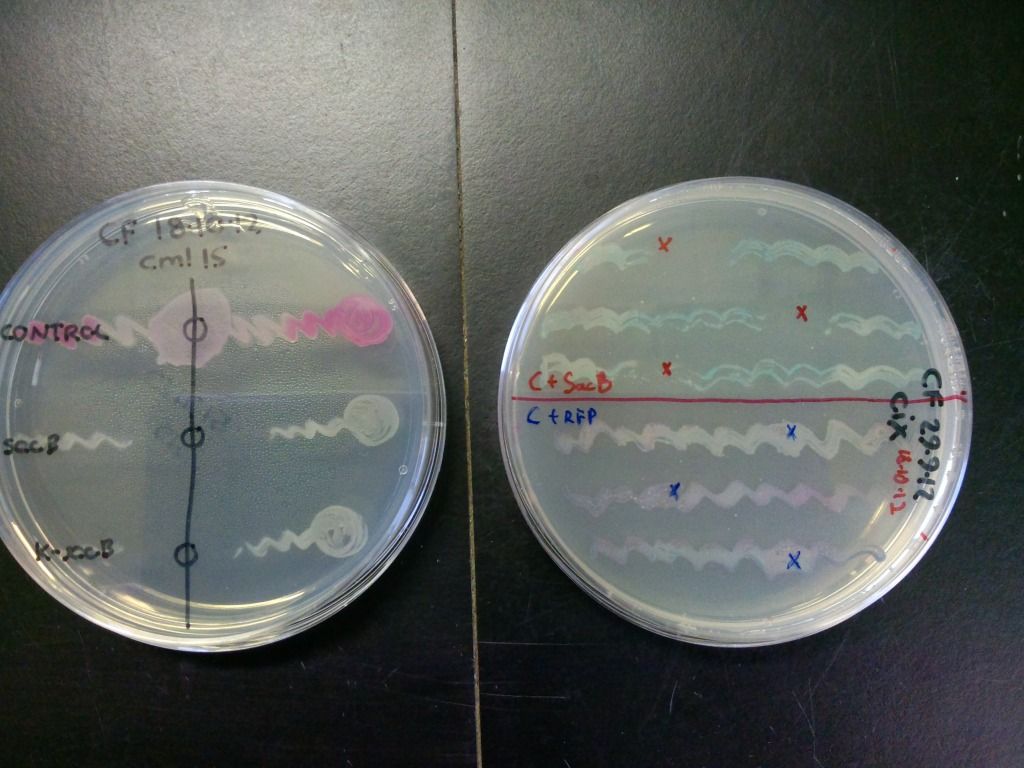
Figure 5: A quick test of sacB and control’s ability to grow in the presence of sucrose. The zone of clearance indicates that the cells containing sacB are inhibited by sucrose while the controls are unaffected. The control on the plate on the right is purple due to both expressing the RFP gene and turning blue because of the X-gal on the plate.
The bottom streak on plate on the left in Figure 5 contains cells that have the selection-counterselection cassette so this test shows that the counterselection component of the cassette is working.
We also tested the selection component of this cassette by streaking the cells onto plates containing both chloramphenicol and kanamycin and incubated overnight. These results can be seen in Figure 6.
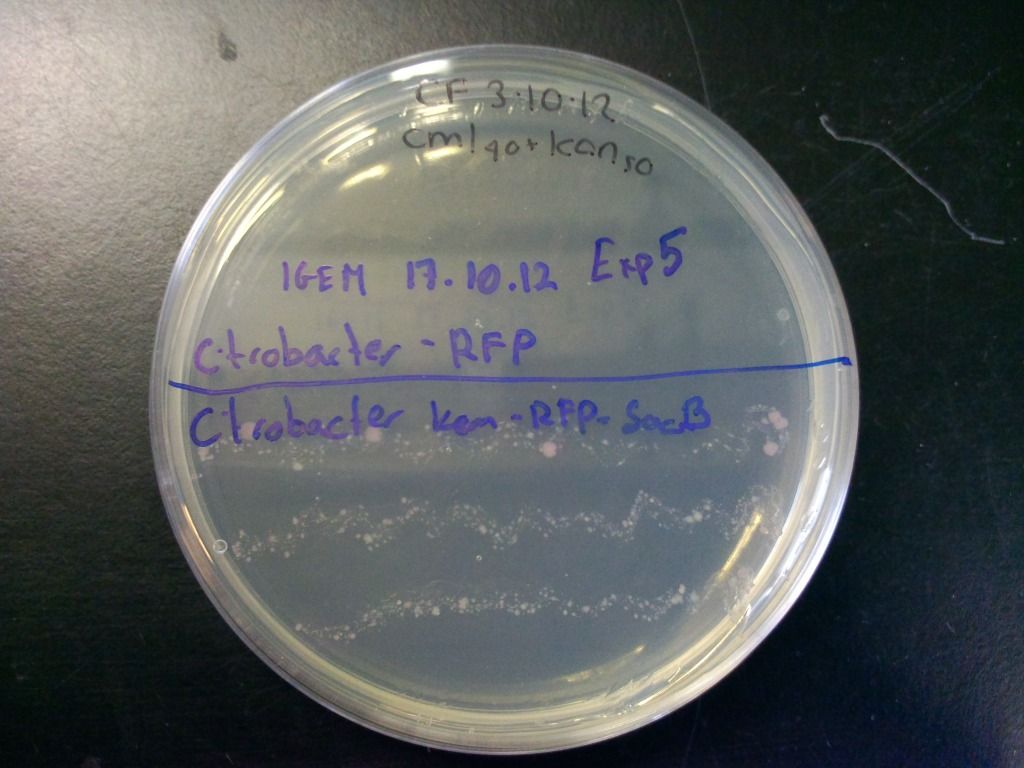
Figure 6: Plate showing that the kanamycin resistance component of the selection-counterselection cassette works. The control did not grow at all, even after 4 days, as it was inhibited by the kanamycin, while the cells containing the cassette show growth. Growth is weaker because we have found that using the concentration of chloramphenicol we normally use (cml40) in combination with the sacB in Citrobacter freundii causes slower growth, presumably because levansucrase expression is unregulated in these bacteria which puts a strain on the cell.
Sugar-dependent selection-counterselection cassette concept
The reason we did not use our sucrose hydrolase gene for the selection component of the selection-counterselection cassette was because both it and levansucrase depend on the same substrate – sucrose – for their function, so this would have not yielded good results.
We have found that Citrobacter freundii can use sucrose as a sole carbon source without the need for the sucrose hydrolase gene – this means that if we want to develop a non-antibiotic resistance-dependent selection-counterselection cassette for this organism, we need to use a different sugar. One sugar that, according to our findings, Citrobacter freundii cannot use is xylitol – see our ‘Xylitol dehydrogenase’ section for more details on this. We could therefore combine this gene with the levansucrase gene to obtain an antibiotic resistance-independent selection-counterselection cassette that dependent entirely on sugars.
Levans
We have found that under sublethal doses of sucrose, the E. coli cells start to secrete a lot of gloopy substance, which we believe to be levans (Figure 7), the fructose polymers formed because of the levansucrase activity.
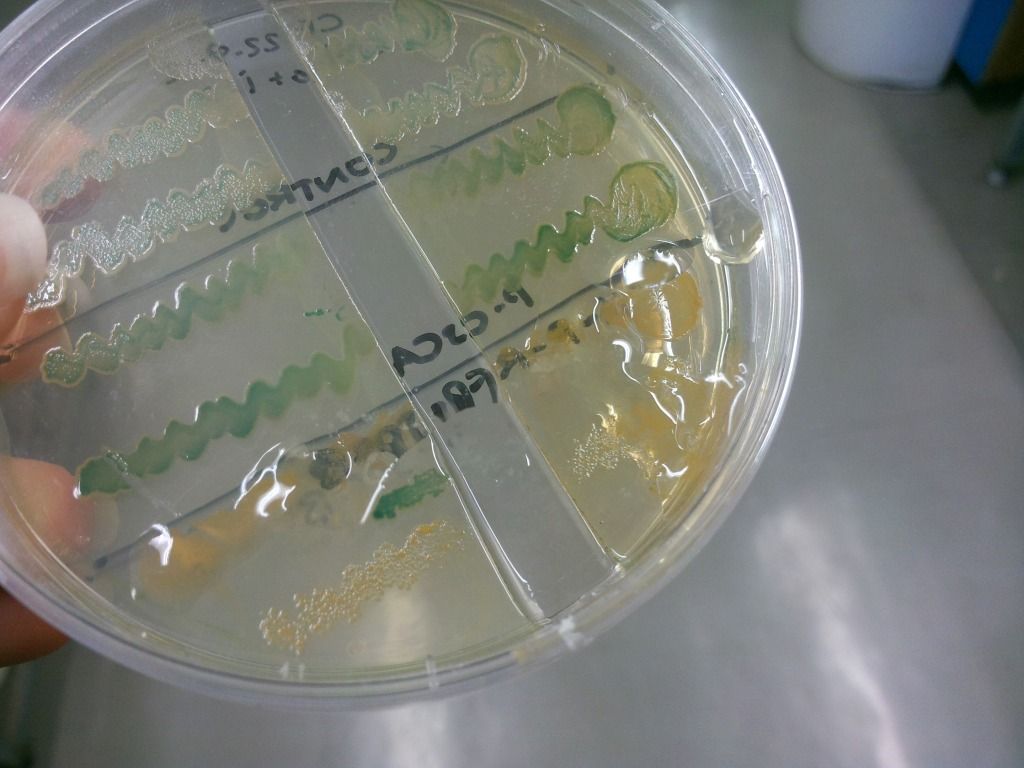
Figure 7: Secretion of levans outside of the cell – it is only present on the lower two colonies but is absent in the two controls above.
We find this important to mention, as levans have got several applications in various fields (Kang et al., 2009).
- Food
- has prebiotic effects
- provides dietary fibres
- reduces serum cholesterol levels
- it can be used as a food additive in the following ways: a stabilizer, an emulsifier, a formulation aid, surface-finishing agent, an encapsulating agent, and a carrier of flavours and fragrances
- Pharmaceutical industry
- can be used as a coating material for drugs
- has anti-tumour properties in vitro
- can be used as a blood plasma volume expander
- has anti-diabetic effects (Dahech et al., 2011)
- levan derivatives are shown to be anti-AIDS agents
- Cosmetics
- good as a cell-proliferating agent
- skin moisturising agent
- reduces skin irritation
- Industry
- it can be used in a two-phase liquid good for the separation of biological samples
- it can be used an environmentally friendly adhesive
- it can be used as a temporary adhesive in water-soluble form
- it can be used as a water-resistant adhesive for a long period of time in cross-linked form
- it can form a water-resistant film
- it acts as a cryoprotectant for the preservation of animal cells and fish
Conclusions:
- We prepared a plac-RFP-SacB construct which can be used for assessing counter-selection efficiency. (BBa_K917002)
- We placed sacB under the lac promoter which allows IPTG dependent control rather than kanamycin dependent control and IPTG concentration-dependent control of the levels of selection.
- We added RFP to allow distinguishing loss of the counter-selection cassette form loss of SacB fuction.
- We prepared and characterized a Kan-plac-RFP-sacB selection-counterselection cassette in both E. coli and Citrobacter freundii (BBa_K917010)
Methods (expand)
Inserting gene into a biobrick vecor: Cloning a PCR product into a biobrick vector protocol on OpenWetWare (http://openwetware.org/wiki/Cfrench:bbcloning) however NEB buffers were used.
DNA gel preparation: Analysing DNA by gel electrophoresis protocol on OpanWetWare (http://openwetware.org/wiki/Cfrench:AGE) however 0.5*TAE rather than 1*TAE was used.
Colony PCR screen: Screening colonies by PCR protocol on OpenWetWare http://openwetware.org/wiki/Cfrench:PCRScreening
Transformations: Preparing and using compenent E.coli cells protocol on OpenWetWare (http://openwetware.org/wiki/Cfrench:compcellprep1)
PCR reactions : Cloning parts by PCR with Kod polymerase protocol on OpenWetWare (http://openwetware.org/wiki/Cfrench:KodPCR)
Minipreps : Plasmid DNA minipreps from Escerichia coli JM109 and similar strains protocol on OpenWetWare (http://openwetware.org/wiki/Cfrench:minipreps1)
Digests to linearise the DNA frangment/determine size of insert: Analytical restriction digests protocol on OpenWetWare (http://openwetware.org/wiki/Cfrench:restriction1)
DNA purification: Purifying a PCR product from solution protocol on OpenWetWare (http://openwetware.org/wiki/Cfrench:DNAPurification1) however 165 ul NaI, 5 ul glass beads,180 ul wash buffer and 10 ul EB were used.
DNA preparation for sequencing: 2.5 ul miniprepped DNA, 2 ul water and 1 ul forward primer ( specific for biobrick prefix) or reverse primer (specific for biobrick suffix)
Nitroreductase activity assay: Overnight liquid cultures of nitroreductase strains were centrifuged at 10000 rpm for 5 mins to pellet the cells. The cells were then resuspended in 250 ul PBS and 1 ul DTT to ensure that cellular proteins are not oxidized. The solution was sonicated 6* (10 s sonication+20 s rest). The supernatant was separated from the pellet by centrifugation and used for the NADH-dependent nitroreductase activity assay.
To assess background activity NADH (5 ul) and bacterial supernatant (5 ul) were added to 0.8 ml PBS and mixed. OD340 was measured for 1 minute. DNBA(5 ul) was added to the same cuvette to start the reaction and change in OD340 was monitored for 1 minute. DMSO(5 ul) was used a control (DNBA is dissolved in DMSO)
The protein concentration of each of the supernants was estimated by by Bradford protein assay using the Pierce reagent protocol on OpenWetWare(http://openwetware.org/wiki/Cfrench:ProteinAssay)
Close methods.
French, C., & Kowal, M. (2010, 09 24). B. subtilis levansucrase. Lethal to E.coli in presence of sucrose. Retrieved 2012, from Registry of standard biological parts: http://partsregistry.org/Part:BBa_K322921
Gay, P., Coq, D. l., Strinmetz, M., Ferrari, E., & Hoch, J. A. (1983). Cloning Structural Gene SacB, which Codes for Exoenzyme Levansucrase of Bacillus subtilis: Expression of the Gene in Esherichia coli. Journal of Bacteriology , 1424-1431.
Jahreis, K., Bentler, L., Bockmann, J., Hans, S., Meyer, A., Siepelmeyer, J., et al. (2002). Adaptation of sucrose metabolism in the Escherichia coli Wild-Type Strain EC31132. Journal of Bacteriology, 5307-5316.
Keuning, S., Janssen, D. B., & Witholt, B. (1985). Purification and Characterisation of Hyrdrolytic Haloalkane Dehalogenase from Xanthobacter autotrophicus GJ10. Journal of Bacteriology, 635-639.
Naested, H., Fennema, M., Hao, L., Andersen, M., Janssen, D. B., & Mundy, J. (1999). A bacterial haloalkane dehalogenase gene as a negative selectable marker in Arabidopsis. The Plant Journal, 571-576.
Nicklin, C. E., & Bruce, N. C. (1998). Aerobic degradation of 2,4,6-Trinitrotoluene by Enterobacter cloaceae PB2 and by Pentaerythritol tetranitrate reductase. Applied and environmental microbiology , 2864-2868.
Nillius, D., Muller, J., & Muller, N. (2011). Nitroreductase (GlNR1) increases susceptibility of Giardia lamblia and Escherichia coli to nitro drugs. Journal of antimicrobial chemotherapy, 1029-1035.
Kang et al. (2009). "Levan: Applications and Perspectives". Microbial Production of Biopolymers and Polymer Precursors. Caister Academic Press
Dahech, I, Belghith, K. S., Hamden, K., Feki, A., Belghith, H. and Mejdoub, H. (2011) Antidiabetic activity of levan polysaccharide in alloxan-induced diabetic rats. International Journal of Biological Macromolecules 49(4):742-746
Close cited works.
 "
"











