Contents |
Miniprep of pSB1C3-RFP
Protocol: Miniprep
The slim tubes can be centrifuged in the machine in front of the "Gel hood", at 4000 rpm for 10 min. The fatter ones, in the E. coli centrifuge by the fridge (the tip can be left inside, since it floats).
Pellets resuspended with RNase containing buffer (Resuspension Buffer R3, from Invitrogen, equivalent to Buffer P1 from Qiagen, in Sowmya's box in the fridge). Note: keep the buffer in ice if you are not bringing it back to the fridge for some minutes.
We then use the QIAGEN QIAprep Spin Miniprep Kit with their [http://www.qiagen.com/literature/render.aspx?id=370 protocol] (page 22) and a microcentrifuge.
Minipreps of the circularized BioBrick plasmid containing the iGEM shipping vector (RFP) were made. However, there was far too much growth in the Falcon tubes, we probably should have put more chloramphenicol.
Nanodrop of the minipreps
Protocol: DNA Concentration Measurement
- Take a 6 µl aliquote of the DNA and put back the main DNA tube in the fridge.
- Go to the room by the E.Coli lab (LBTM, not on Friday morning!) with:
- The 6 µl aliquote
- A 10 µl pipet
- Optionally, the buffer you used for DNA elution (there might be some next to the machine).
- The machine is the NanoDrop Spectrophotometer.
- On the computer, click on "Nucleic Acid".
- Put a 2 µl drop of (nuclease-free) water on the machine's tip as you are asked to and measure.
- Clean tips (both sides) with a quarter of tissue.
- Add 2 µl of the buffer you use and click on "Blank".
- Clean tips (both sides).
- Add 2 µl of your DNA sample and click "Measure".
- Clean tips (both sides) with a tissue.
- Take 2 measurements per sample (for averaging).
- Print the report when you are done
- Click on exit.
The important numbers are:
- 260/280 ratio, must be > 1.8
- 260/230 ratio, must be > 2 (too big, > 2.5? , might mean too much salts)
- Of course the DNA concentration.
The Nanodrop showed dreadfully low concentrations. It was decided that we would redo a culture with a higher chloramphenicol concentration.
New overnight cultures
Protocol: Prepare Plasmid Extraction (culture for Miniprep)
- Select and number colonies on the plates.
- Prepare tubes of LB medium with the correct quantity of antibiotics (100 µg/ml for Amp, Spc or chloramphenicol).
- Amp can be found in the -20ºC freezer at Ecoli, labeled as "stock". It is 100 µg/µl, or 1000x.
- The tubes to be used are the 14 ml round bottom found in front of the iGEM drawers (Falcon). Culture with cap in the first step (loose) and close to the second step after culture.
- Touch each colony with a clean pipette tip and put it in a tube.
- Put in incubator.
Overnight cultures of pSB1C3-RFP, pSB1C3-LovTAP and pCEP4-HA-RO were made for miniprepping. The first two were cultured on chloramphenicol with a higher concentration. We also made control cultures to see if ampicillin-resistant bacteria would grow on chloramphenicol, and the opposite.
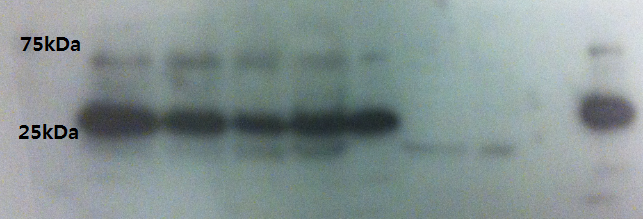
Western Blot result from 5.sep.12
Protocol: Western Blot
Gel Ingredients (choose percentage according to the size of the protein)
| 4-40 kDA | 20% |
| 12-45 kDA | 15% |
| 10-70 kDA | 12.5% |
| 15-100 kDA | 10% |
| 25-200 kDA | 8% |
| Separating gel | |
| Gel percentage | 7.5 % |
| 30% Polyacrylamide | 10 mL |
| 1.5M Tris (pH 8.8) | 10 mL |
| 10% Ammonium persulfate | 0.4 mL |
| 10% SDS | 0.4 mL |
| TEMED | 0.038 mL |
| H2O | 19.2 mL |
| Total volume | 40 mL |
| Stacking gel | |
| Gel percentage | 5 % |
| 30% Polyacrylamide | 1.36 mL |
| 1M Tris (pH 6.8) | 1 mL |
| 10% Ammonium persulfate | 0.08 mL |
| 10% SDS | 0.08 mL |
| TEMED | 0.008 mL |
| H2O | 5.44 mL |
| Total volume | 8 mL |
Preparing Protein Samples
1. Centrifuge around 5 million cells (of any volume) at 2,500 rpm for 10 min.
2. Discard the supernatant with a vacuum pump.
3. Resuspend the cell pellet with 1x PBS and centrifuge it at 2,500 rpm for 10 min.
4. Discard the supernatant with a vacuum pump.
5. Add appropriate amount of lysis buffer depending on the pellet size (for a 20 mg pellet, 150 µl of IP lysis buffer).
6. Keep the lysed sample on ice for 10 min - flick every 3 minutes.
7. Add 3x SDS lysis buffer (for a 20 mg pellet, 75 µl).
8. Incubate the sample for 5 minutes at 95 degrees, to denature proteins.
Preparing loading samples
1. Load the ladder (7 µl is the recommended volume).
2. Complete sample volume to 50 µl.
3. Load the samples.
I. SDS Gel electrophoresis
1. Prepare the separating and stacking gel solutions without APS and TEMED.
2. Add APS and TEMED to the separating gel solution only when the SDS kit is ready to be used, they are time-sensitive. Move the solution inside of the setup. Add some distilled water on top of it.
3. After 20-30 mins, remove the water and check whether the gel has solidified. Don't move to the next step until it does.
4. Add TEMED to the stacking gel solution, pour it on top of the solidified separating gel.
5. Insert a stack carefully and leave it for 20-30 mins.
6. Take the stack out and fill the kit with SDS loading buffer.
7. Load the samples.
8. Add more loading buffer, set the voltage to 80 Volts. Leave for 1.5 hours.
II. Membrane transfer
1. Prepare a membrane transfer kit.
2. Take the gel out of the SDS kit and put it on the membrane paper.
3. From bottom to top, assemble the components in the following order: 1) Sponge - 2) Blot paper - 3) Membrane - 4) Gel (Pour some M-transfer buffer on the gel) - 5) Blot paper again - 6) Sponge again.
4. Close the sandwich, set the voltage to 20 V. Leave for 30 mins - 1 hour.
5. Discard the gel. Leave the membrane in 5% skim milk with 30ml of TBST buffer (blocking buffer, to achieve the 5%, add 1.5 g of skim milk powder to the buffer) for one hour.
III. Antibody tagging
1. Discard the blocking buffer, leave only 5ml of it. Add primary antibody with a ratio of 1:1000 or 1:2000 (5 µl of antibody in 5 ml of buffer gives 1:1000)
2. Leave the mix overnight at 4 °C.
3. Wash 3 times with 1x TBST (5 minutes on shaker for every wash).
4. Dilute the secondary antibody (for example, goat anti-rabbit antibody) to 1:2000 in 5% skim milk buffer. Add it. Leave at room temperature for 2 hours.
5. Wash 3 times with 1x TBST (5 minutes on shaker for every wash).
6. Reveal the protein bands in the dark room.
Double-check the VP16 antibody detection to see if it's consistent with the result from the previous day.
- Lane 1: Ladder
- Lane 2: CHO cell lysate with VP16 protein 5microG
- Lane 3: CHO cell lysate with VP16 protein 2microG
- Lane 4: CHO cell lysate with VP16 protein 1microG
- Lane 5: CHO cell lysate with VP16 protein 500nanoG
- Lane 6: CHO cell lysate with VP16 protein 100nanoG
- Lane 7: Empty
- Lane 8: Empty
- Lane 9: Empty
- Lane 10: CHO cell lysate with VP16 protein 5microG
It seems quite obvious that our VP16 protein is really around 35kDa size not 25kDa.
Now we are going to WB with transfected CHO cells since we have practiced and checked our antibody detects VP16 and we don't have too many non specific bands with CHO cell lysates.
 "
"