Team:TU Munich/Project/Caffeine
From 2012.igem.org



Contents |
Caffeine
In contrast, caffeine wards of drowsiness and already enjoys great popularity as additive in a multitude of beverages. As hops is essential to the brewing process, omitting is not an option. This makes caffeine a desirable agent to counteract the soporific effect of beer.
Background and principles
At higher doses (1g), caffeine leads to higher pulse rates and hyperactivity, but until that, the beer will already have done its work... Caffeine was shown to decrease the growth of E. Coli and Yeast reversibly as of a concentration of 0.1% by acting as a mutagen (Putrament et al., On the Specificity of Caffeine Effects, MGG, 1972), but previous caffeine synthesis experiments (see below) have only led to a concentration of about 5 µg/g (per g fresh weight of tobacco leaves), so we do not expect to reach critical concentrations. It has already been achieved to produce caffeine in tobacco plants (Uefuji et al., 2005; Yun- Soo Kim et al., 2007), but has never been performed in yeast.
Biosynthesis
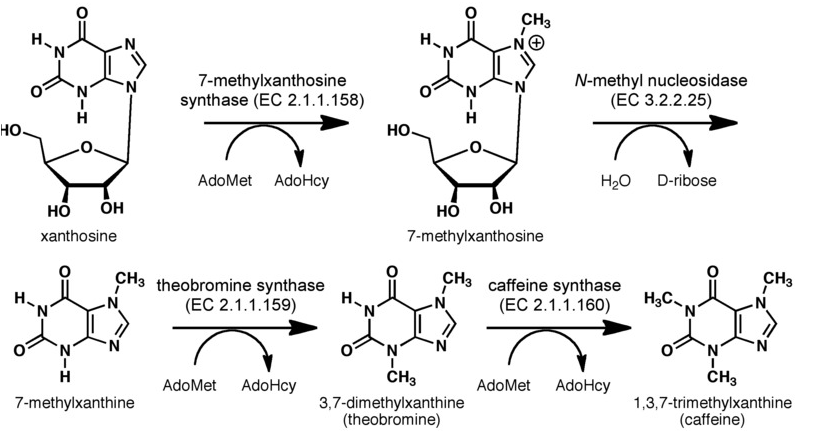
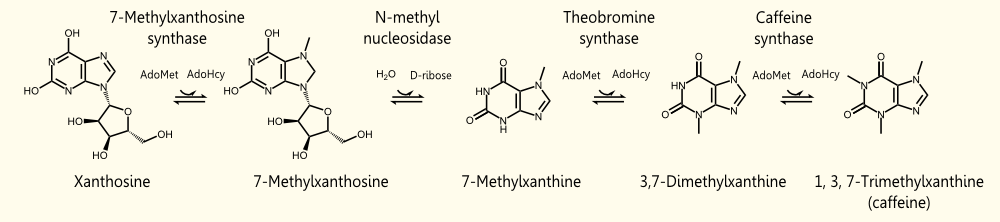
The biosynthetic pathway of caffeine (1,3,7 Trimethylxanthine) starts with xanthosine, which is a natural component of the purine- metabolism of all organism. Necessary for its production are three distinct N- methyl transferases and one nucleosidase, whereupon it has not been totally elucidated whether the nucleosidase reaction is catalyzed by any purine nucleosidase or by the first N- methyl transferase of the reaction cascade shown in the picture (but the latter assumption is favoured (H. Ashihara et al., 2008), because an in vitro synthesis of caffeine with the three N- methyl transferases has already been shown). The enzyme caffeine synthase (last reaction step) can catalyze both, the conversion of 7- methyl xanthine to theobromine and the methylation of theobromine to caffeine. This is true, indeed, but Uefuji et al. (2003) showed, that the affinity to 7- methyl xanthosine is less than one sixth of that of CaMXMT1 (there are two isoformes of CaMXMT), so it is much better to express both enzymes. One can also see the Km values for the required enzymes in this paper - it shows that the substrate affinity decreases continiously towards the endpoint (caffeine), "making the reaction proceed irreversibly and stepwise" (Uefuji et al., 2003, p.377).
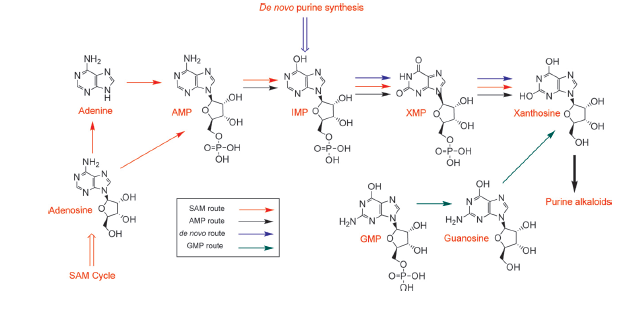
The chemical compound xanthosine is produced via at least four different routes, shown in the picture "xanthosine routes". To improve caffeine production, these pathways could be a possible target for metabolic engineering. Anyway it should be interesting/ necessary for this project to determine the in vivo xanthosine concentration of yeast.
Catabolism
- please delete this heading to ensure unity in all our pages
Caffeine is demethylated to theophylline by 7N-demethylase (main pathway). The decreased rate of this reaction is the reason for accumulating caffeine in the plant. Afterwards, theophylline is degraded to xanthine via 3-methylxanthine and xanthine enters the conventional purine catabolism pathway (degradation to CO2 and NH3) (see H. Ashihara et al., 2008, p. 846). This catabolistic pathway is another possible target for metabolic engineering to increase the amount of caffeine.
The molecular and physiological effects of caffeine
Results
BioBricks
- include here all the informations about the enzymes
Characterization
Gene expression
The expression of the three neccessary enzymes involved in caffeine biosynthesis will be prooved by SDS page and Western Blot analysis, making use of the Strep-tag II and a detection system based on two antibodies: StrepMAB-Classic (from mouse) and an Anti-Mouse-alk. phosphatase conjugate.
Enzyme-function
First of all, the function of the three enzymes will be demonstrated in vitro in a combinated reaction batch, containing all three enzymes. After adding the initial substrate xanthosine, we will detect the synthesized caffeine after organic extraction and HPLC separation (densitometrically).
Function of expression cartridge and membran-permeability of compounds
At first, the fuction of our expression cartridge will be prooved by Western Blot analysis, as mentioned above. To investigate wether both, the initial substrate xanthosine and the final product caffeine are able to penetrate the yeast cell wall, we will apply the substrate xanthosine to a cell suspension, in which the cells contain our created "caffeine synthesis biobrick" with the constitutive promoters Tef1 and Tef2. Caffeine presence will be detected as mentioned above.
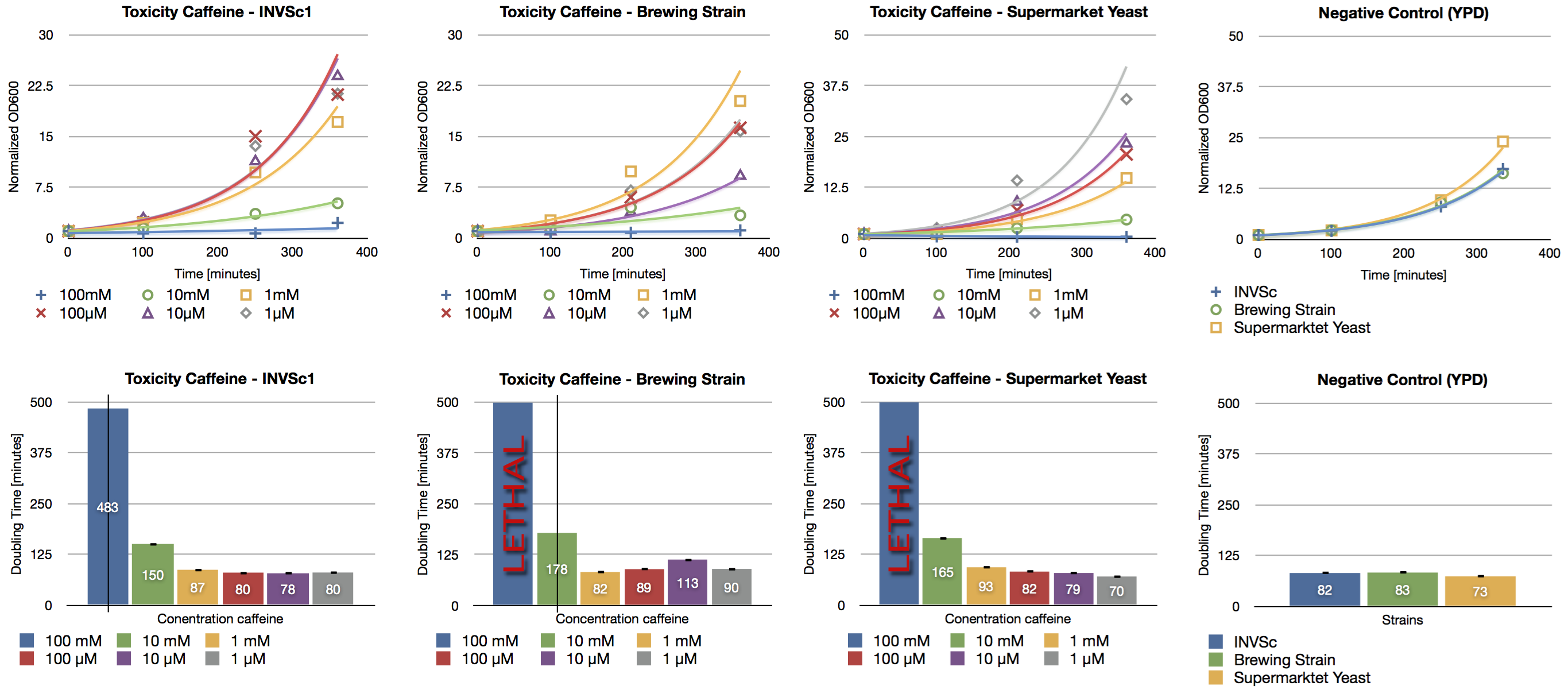
Toxicity Assay
References
- Putrament et al., On the Specificity of Caffeine Effects, MGG, 1972
- H. Uefuji et al., Plant Physiology, 2003, Vol. 132, pp. 372–380
- H. Uefuji et al., Plant Molecular Biology, 2005, Vol. 59, p. 221–227
- H. Ashihara et al., Phytochemistry, 2008, Vol. 69, p. 841–856
- Yun-Soo Kim, Hiroshi Sano, Phytochemistry, 2008, Vol. 69, p. 882–888
- Franco et al., 2012
older blocks
- please integrate the informations into the other blocks and delete the sequences here (copy them into the registry)
Idea
The idea is to perform a heterologous gene expression of the distinct N-methyl transferases required for caffeine biosynthesis. At first, this will be accomplished by cloning each gene in a single, RFC25 compatible pYES2 vector, which will be created by the iGEM- team TU- Munich 2012. Expression will be proofed by western blot analysis, making use of a strep-tagII. Simultaneously, we will construct composite parts, each consisting of coding sequence, Adh1 terminator and a constitutive promoter with appropriate strength. The chosen promoters are Tef2 for CaXMT1 and CaDXMT1 and Tef1 for CaMXMT1. This decission was made with respect to the fact, that the synthesis of caffeine shall be an irreversible reaction. In vivo, this irreversibility is realized by continuously increasing Km values (Km(CaXMT1) < Km(CaMXMT1) < Km(CaDXMT1)); as soon as a certain amount of theobromine is available, the caffeine synthesis can go on. We will support this stepwise reaction- principle by using the strongest promoter for the theobromine synthesis (performed by CaMXMT1), resulting in higher concentrations of this chemical compound and making the last reaction step irreversible. In order to limit the metabolic stress, we will use the weaker Tef2 promoter for the first and last enzymes CaXMT1 and CaDXMT1, although it could theoretically lead to higher amounts of caffeine, when using a stronger initial promoter. Finally, we will fuse all these three composite parts and create the "caffeine synthesis device", which can be used for genome integration in yeasts.
The research groups which accomplished caffeine production in transgenic tobacco plants used the following three genes:
- CaXMT1 (AB048793)
- UniProt entry: Q9AVK0
- E.C.: 2.1.1.158
- PDB: 3D- Structure: http://www.pdb.org/pdb/explore/explore.do?structureId=2eg5 (Structure of C. canephora- CaXMT1)
- Complete mRNA- sequence: http://www.ncbi.nlm.nih.gov/nuccore/AB048793 CaXMT1 (coffea arabica)
- Coding sequence: bp 45 - 1163 ==> total length: 1119 bp
- Problematic restriction sites: EcoR1 at bp 104- 109
- CaMXMT1 (AB048794)
- UniProt entry: Q9AVJ9
- 2.1.1.159
- This gene is eventually not essential, for CaDXMT1 is able to catalyze this reaction, too.
- Complete mRNA- sequence: http://www.ncbi.nlm.nih.gov/nuccore/AB048794 CaMXMT1 (coffea arabica)
- Coding sequence: bp 32 - 1168 ==> total length: 1137 bp
- Problematic restriction sites: EcoR1 at bp 722- 728
- CaDXMT1 (AB084125)
- UniProt entry: Q8H0D2
- 2.1.1.160
- PDB: 3D- Structure: http://www.pdb.org/pdb/explore/explore.do?structureId=2efj (Structure of C. canephora- CaDXMT1)
- Complete mRNA- sequence: http://www.ncbi.nlm.nih.gov/nuccore/AB084125 CaDXMT1 (coffea arabica)
- Coding sequence: bp 1- 1155 ==> total length: 1155 bp
- Problematic restriction sites: EcoR1 at bp 691- 697 and at bp 705- 711
All mentionend methyltransferases use SAM als methyl- donor and are located in the cytoplasm of the plants. Furthermore they exist as homodimers, being also able to form heterodimers with each other (see http://www.brenda-enzymes.info BRENDA, also for further characteristics). The temperature and pH optimum of all three enzymes is quite similar between 20°C - 37°C and 7,5 - 8,5, respectively (beer brewing: slightly acid).
General remarks and issues
Analytical Methods
- Detection and quantification of the produced caffeine can be performed by the use of HPLC. Uefuji et al., 2005; describe those and other relevant methods in this context.
- Proof of the gene- expression (of the necessary methyl- transferases) could be accomplished by standard methods (Western Blot, see also Uefuji et al., 2005; and Yun- Soo Kim and Hiroshi Sano, 2007;)
Gene Expression
To increase gene expression, we are taking care about the following aspects:
- N-end rule
- codon optimization
- yeast consensus sequence for improved ribosome binding
- use of adequate promoters
Since all enzymes are localized in the cytoplasm, they should all be soluble.
Ordered Sequences
The sequences were ordered and synthesized by GeneArt and constructed as follows:
- the 5' UTR and 3' UTR of the sequences above were removed
- the yeast consensus sequence for improved ribosome binding (TACACA) was added 5' of the start codon ATG
- according to N- end rule and the yeast consensus sequence for improved ribosome binding, the first triplet after ATG (GAG) was exchanged with TCT (serine), to optimize both, protein stability and mRNA translation. This decision was made after proofing the 3D- structure of the enzyme CaDXMT1. Due to the fact, that the the first two residues of the amino acid sequence are not shown in the crystalized structure (probably because of high flexibility), Prof. Skerra said we can risk the exchange of this amino acid, for it is probably not that necessary for the uptake of the ligands (uniprot entry further shows, that it is not immediately involved in ligand binding in one of the three enzymes). Because of the high similarity of the enzyme- sequences, we also exchanged this amino acid in the enzyme CaMXMT1, although here is no 3D- structure available
- we added a c- terminal strep-tag for purification
- the remaining coding sequence was extended with the standard RFC10 prefix and suffix, respectively
- at last we made an optimization of the coding sequences with respect to the codon usage and mRNA structures (online tool of a gene- synthesis company)
- annotated sequences:
CaXMT1
10 20 30 40 50 60 70 80 90 100
* * * * * * * * * *
1 gaattcgcggccgcttctagagtacacaatgtctttacaagaagtcttgagaatgaacggtggtgaaggtgatacttcttacgctaaaaactccgcctac 100
cttaagcgccggcgaagatctcatgtgttacagaaatgttcttcagaactcttacttgccaccacttccactatgaagaatgcgatttttgaggcggatg
M S L Q E V L R M N G G E G D T S Y A K N S A Y
1
>>>>>>>>>>>>>>>>>>>>>> >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
RFC10 Prefix CaXMT1_Strep-tag
>>>>>>>>>>>>
RBS Yeast Consensus Sequence
110 120 130 140 150 160 170 180 190 200
* * * * * * * * * *
101 aatcaattggttttggctaaagttaagccagtcttggaacaatgcgtcagagaattattgagagctaacttgccaaacatcaacaagtgcattaaggttg 200
ttagttaaccaaaaccgatttcaattcggtcagaaccttgttacgcagtctcttaataactctcgattgaacggtttgtagttgttcacgtaattccaac
N Q L V L A K V K P V L E Q C V R E L L R A N L P N I N K C I K V A
25
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
210 220 230 240 250 260 270 280 290 300
* * * * * * * * * *
201 ctgatttgggttgtgcttctggtccaaatactttgttgactgttagagacatcgtccaatccattgataaggttggtcaagaaaagaagaacgaattgga 300
gactaaacccaacacgaagaccaggtttatgaaacaactgacaatctctgtagcaggttaggtaactattccaaccagttcttttcttcttgcttaacct
D L G C A S G P N T L L T V R D I V Q S I D K V G Q E K K N E L E
59
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
310 320 330 340 350 360 370 380 390 400
* * * * * * * * * *
301 aagaccaaccatccaaattttcttgaacgacttgttcccaaacgacttcaactctgtttttaagttgttgccatccttctacagaaagttggaaaaagaa 400
ttctggttggtaggtttaaaagaacttgctgaacaagggtttgctgaagttgagacaaaaattcaacaacggtaggaagatgtctttcaacctttttctt
R P T I Q I F L N D L F P N D F N S V F K L L P S F Y R K L E K E
92
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
410 420 430 440 450 460 470 480 490 500
* * * * * * * * * *
401 aacggtagaaagatcggttcctgtttgattggtgctatgccaggttctttctactccagattatttcctgaagaatccatgcatttcttgcactcttgtt 500
ttgccatctttctagccaaggacaaactaaccacgatacggtccaagaaagatgaggtctaataaaggacttcttaggtacgtaaagaacgtgagaacaa
N G R K I G S C L I G A M P G S F Y S R L F P E E S M H F L H S C Y
125
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
510 520 530 540 550 560 570 580 590 600
* * * * * * * * * *
501 attgcttgcaatggttgtctcaagttccatctggtttggttactgaattgggtatttctaccaacaagggttccatctactcttctaaagcttcaagatt 600
taacgaacgttaccaacagagttcaaggtagaccaaaccaatgacttaacccataaagatggttgttcccaaggtagatgagaagatttcgaagttctaa
C L Q W L S Q V P S G L V T E L G I S T N K G S I Y S S K A S R L
159
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
610 620 630 640 650 660 670 680 690 700
* * * * * * * * * *
601 gccagttcaaaaggcctacttggatcaattcactaaggatttcaccacctttttgagaatccactccgaagaattattctcccacggtagaatgttgttg 700
cggtcaagttttccggatgaacctagttaagtgattcctaaagtggtggaaaaactcttaggtgaggcttcttaataagagggtgccatcttacaacaac
P V Q K A Y L D Q F T K D F T T F L R I H S E E L F S H G R M L L
192
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
710 720 730 740 750 760 770 780 790 800
* * * * * * * * * *
701 acctgtatatgtaagggtgttgaattggatgctagaaacgccattgatttgttggaaatggctatcaacgatttggttgttgaaggtcacttagaagaag 800
tggacatatacattcccacaacttaacctacgatctttgcggtaactaaacaacctttaccgatagttgctaaaccaacaacttccagtgaatcttcttc
T C I C K G V E L D A R N A I D L L E M A I N D L V V E G H L E E E
225
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
810 820 830 840 850 860 870 880 890 900
* * * * * * * * * *
801 aaaagttggactctttcaacttgccagtttacattccatctgccgaagaagttaagtgcatcgttgaagaagaaggttccttcgaaatcttgtacttgga 900
ttttcaacctgagaaagttgaacggtcaaatgtaaggtagacggcttcttcaattcacgtagcaacttcttcttccaaggaagctttagaacatgaacct
K L D S F N L P V Y I P S A E E V K C I V E E E G S F E I L Y L E
259
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
910 920 930 940 950 960 970 980 990 1000
* * * * * * * * * *
901 aactttcaaggtcttgtacgatgccggtttctctattgatgatgaacatattaaggccgaatacgttgcctcttctgttagagctgtttacgaacctatt 1000
ttgaaagttccagaacatgctacggccaaagagataactactacttgtataattccggcttatgcaacggagaagacaatctcgacaaatgcttggataa
T F K V L Y D A G F S I D D E H I K A E Y V A S S V R A V Y E P I
292
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
1010 1020 1030 1040 1050 1060 1070 1080 1090 1100
* * * * * * * * * *
1001 ttggcttctcatttcggtgaagccattatcccagatatctttcatagatttgctaagcacgctgctaaggttttgccattgggtaaaggtttttacaaca 1100
aaccgaagagtaaagccacttcggtaatagggtctatagaaagtatctaaacgattcgtgcgacgattccaaaacggtaacccatttccaaaaatgttgt
L A S H F G E A I I P D I F H R F A K H A A K V L P L G K G F Y N N
325
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
1110 1120 1130 1140 1150 1160 1170 1180 1190 1200
* * * * * * * * * *
1101 acttgatcatctccttggccaaaaagccagaaaagtctgatgtttctgcttggtcacatccacaattcgaaaagtgatgatactagtagcggccgctgca 1200
tgaactagtagaggaaccggtttttcggtcttttcagactacaaagacgaaccagtgtaggtgttaagcttttcactactatgatcatcgccggcgacgt
>>>>>>>>>>>>>>>>>>>>
RFC10 Suffix
L I I S L A K K P E K S D V S A W S H P Q F E K * *
359
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaXMT1_Strep-tag
1201 g 1201
c
>
RFC10 Suffix
Theoretical molecular weight: 42998,4 Da (ProtParam)
Probable posttranslational modifications:
- acetylation at serine (second amino acid)
- O-GlcNAc modifikation at several positions
- phosphorylation at several positions
(see http://2d.bjmu.edu.cn/show2d/Proteomics%20tools.asp)
CaMXMT1
10 20 30 40 50 60 70 80 90 100
* * * * * * * * * *
1 gaattcgcggccgcttctagagtacacaatgtctttacaagaagtcttgcacatgaacgaaggtgaaggtgatacttcttacgctaagaatgcttcttac 100
cttaagcgccggcgaagatctcatgtgttacagaaatgttcttcagaacgtgtacttgcttccacttccactatgaagaatgcgattcttacgaagaatg
M S L Q E V L H M N E G E G D T S Y A K N A S Y
1
>>>>>>>>>>>>>>>>>>>>>> >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
RFC10 Prefix CaMXMT1_Strep-tag
>>>>>>>>>>>>
RBS Yeast Consensus Sequence
110 120 130 140 150 160 170 180 190 200
* * * * * * * * * *
101 aacttggctttggctaaggttaagccattcttggaacaatgcatcagagaattattgagagccaacttgccaaacatcaacaagtgtattaaggttgctg 200
ttgaaccgaaaccgattccaattcggtaagaaccttgttacgtagtctcttaataactctcggttgaacggtttgtagttgttcacataattccaacgac
N L A L A K V K P F L E Q C I R E L L R A N L P N I N K C I K V A D
25
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
210 220 230 240 250 260 270 280 290 300
* * * * * * * * * *
201 atttgggttgtgcttctggtccaaatactttgttgactgttagagacatcgtccaatccattgataaggttggtcaagaagaaaagaacgaattggaaag 300
taaacccaacacgaagaccaggtttatgaaacaactgacaatctctgtagcaggttaggtaactattccaaccagttcttcttttcttgcttaacctttc
L G C A S G P N T L L T V R D I V Q S I D K V G Q E E K N E L E R
59
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
310 320 330 340 350 360 370 380 390 400
* * * * * * * * * *
301 accaaccatccaaattttcttgaacgacttgttccaaaacgacttcaactccgtttttaagttgttgccatccttctacagaaagttggaaaaagaaaac 400
tggttggtaggtttaaaagaacttgctgaacaaggttttgctgaagttgaggcaaaaattcaacaacggtaggaagatgtctttcaacctttttcttttg
P T I Q I F L N D L F Q N D F N S V F K L L P S F Y R K L E K E N
92
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
410 420 430 440 450 460 470 480 490 500
* * * * * * * * * *
401 ggtagaaagatcggttcctgcttgatttctgctatgccaggttctttttacggtagattattccctgaagaatccatgcatttcttgcactcttgttact 500
ccatctttctagccaaggacgaactaaagacgatacggtccaagaaaaatgccatctaataagggacttcttaggtacgtaaagaacgtgagaacaatga
G R K I G S C L I S A M P G S F Y G R L F P E E S M H F L H S C Y S
125
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
510 520 530 540 550 560 570 580 590 600
* * * * * * * * * *
501 ctgttcactggttgtctcaagttccatctggtttggttattgaattgggtattggtgctaacaagggttccatctattcttctaaaggttgtagaccacc 600
gacaagtgaccaacagagttcaaggtagaccaaaccaataacttaacccataaccacgattgttcccaaggtagataagaagatttccaacatctggtgg
V H W L S Q V P S G L V I E L G I G A N K G S I Y S S K G C R P P
159
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
610 620 630 640 650 660 670 680 690 700
* * * * * * * * * *
601 agttcaaaaggcttacttggatcaattcactaaggacttcaccactttcttgagaatccactccaaagaattattctccagaggtagaatgttgttgacc 700
tcaagttttccgaatgaacctagttaagtgattcctgaagtggtgaaagaactcttaggtgaggtttcttaataagaggtctccatcttacaacaactgg
V Q K A Y L D Q F T K D F T T F L R I H S K E L F S R G R M L L T
192
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
710 720 730 740 750 760 770 780 790 800
* * * * * * * * * *
701 tgtatctgtaaggttgacgaatttgatgaacctaacccattggatttgttggatatggccattaacgatttgatcgtcgaaggtttgttggaagaagaaa 800
acatagacattccaactgcttaaactacttggattgggtaacctaaacaacctataccggtaattgctaaactagcagcttccaaacaaccttcttcttt
C I C K V D E F D E P N P L D L L D M A I N D L I V E G L L E E E K
225
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
810 820 830 840 850 860 870 880 890 900
* * * * * * * * * *
801 agttggactccttcaacattccattctttactccatctgccgaagaagttaagtgcatcgttgaagaagaaggttcttgcgaaatcttgtacttggaaac 900
tcaacctgaggaagttgtaaggtaagaaatgaggtagacggcttcttcaattcacgtagcaacttcttcttccaagaacgctttagaacatgaacctttg
L D S F N I P F F T P S A E E V K C I V E E E G S C E I L Y L E T
259
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
910 920 930 940 950 960 970 980 990 1000
* * * * * * * * * *
901 tttcaaggctcattacgatgctgccttctctattgatgatgattacccagttagatcccacgaacaaatcaaagctgaatacgttgcctccttgatcaga 1000
aaagttccgagtaatgctacgacggaagagataactactactaatgggtcaatctagggtgcttgtttagtttcgacttatgcaacggaggaactagtct
F K A H Y D A A F S I D D D Y P V R S H E Q I K A E Y V A S L I R
292
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
1010 1020 1030 1040 1050 1060 1070 1080 1090 1100
* * * * * * * * * *
1001 tctgtttacgaacctattttggcctctcatttcggtgaagctattatgccagatttgtttcacagattggctaaacatgctgctaaggttttacacatgg 1100
agacaaatgcttggataaaaccggagagtaaagccacttcgataatacggtctaaacaaagtgtctaaccgatttgtacgacgattccaaaatgtgtacc
S V Y E P I L A S H F G E A I M P D L F H R L A K H A A K V L H M G
325
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
1110 1120 1130 1140 1150 1160 1170 1180 1190 1200
* * * * * * * * * *
1101 gtaagggttgttacaacaacttgattatctccttggccaaaaagccagaaaagtctgatgtttctgcttggtcacatccacaattcgaaaagtgatgata 1200
cattcccaacaatgttgttgaactaatagaggaaccggtttttcggtcttttcagactacaaagacgaaccagtgtaggtgttaagcttttcactactat
>>
RFC10 Suffix
K G C Y N N L I I S L A K K P E K S D V S A W S H P Q F E K * *
359
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaMXMT1_Strep-tag
1210
*
1201 ctagtagcggccgctgcag 1219
gatcatcgccggcgacgtc
>>>>>>>>>>>>>>>>>>>
RFC10 Suffix
Theoretical molecular weight: 43903,3 Da (ProtParam)
Probable posttranslational modifications:
- acetylation at serine (second amino acid)
- O-GlcNAc modifikation at several positions
- phosphorylation at several positions
(see http://2d.bjmu.edu.cn/show2d/Proteomics%20tools.asp)
CaDXMT1
10 20 30 40 50 60 70 80 90 100
* * * * * * * * * *
1 gaattcgcggccgcttctagagtacacaatgtctttacaagaagtcttgcatatgaacggtggtgaaggtgatacttcttacgctaagaactctttctac 100
cttaagcgccggcgaagatctcatgtgttacagaaatgttcttcagaacgtatacttgccaccacttccactatgaagaatgcgattcttgagaaagatg
M S L Q E V L H M N G G E G D T S Y A K N S F Y
1
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
>>>>>>>>>>>>>>>>>>>>>>
RFC10 Prefix
>>>>>>>>>>>>
RBS Yeast Consensus Sequence
110 120 130 140 150 160 170 180 190 200
* * * * * * * * * *
101 aacttgttcttgatcagagtcaagccaatcttggaacaatgcatccaagaattattgagagccaacttgccaaacatcaacaagtgtattaaggttgctg 200
ttgaacaagaactagtctcagttcggttagaaccttgttacgtaggttcttaataactctcggttgaacggtttgtagttgttcacataattccaacgac
N L F L I R V K P I L E Q C I Q E L L R A N L P N I N K C I K V A D
25
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
210 220 230 240 250 260 270 280 290 300
* * * * * * * * * *
201 atttgggttgtgcttctggtccaaatactttgttgactgttagagacatcgtccaatccattgataaggttggtcaagaaaagaagaacgaattggaaag 300
taaacccaacacgaagaccaggtttatgaaacaactgacaatctctgtagcaggttaggtaactattccaaccagttcttttcttcttgcttaacctttc
L G C A S G P N T L L T V R D I V Q S I D K V G Q E K K N E L E R
59
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
310 320 330 340 350 360 370 380 390 400
* * * * * * * * * *
301 accaaccatccaaattttcttgaacgacttgttccaaaacgacttcaactccgtttttaagtccttgccatccttctacagaaagttggaaaaagaaaac 400
tggttggtaggtttaaaagaacttgctgaacaaggttttgctgaagttgaggcaaaaattcaggaacggtaggaagatgtctttcaacctttttcttttg
P T I Q I F L N D L F Q N D F N S V F K S L P S F Y R K L E K E N
92
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
410 420 430 440 450 460 470 480 490 500
* * * * * * * * * *
401 ggtagaaagatcggttcctgtttgattggtgctatgccaggttctttttacggtagattattccctgaagaatccatgcatttcttgcattcttgttact 500
ccatctttctagccaaggacaaactaaccacgatacggtccaagaaaaatgccatctaataagggacttcttaggtacgtaaagaacgtaagaacaatga
G R K I G S C L I G A M P G S F Y G R L F P E E S M H F L H S C Y C
125
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
510 520 530 540 550 560 570 580 590 600
* * * * * * * * * *
501 gcttgcactggttgtctcaagttccatctggtttggttactgaattgggtatttctgctaacaagggttgcatctactcttctaaagcttcaagaccacc 600
cgaacgtgaccaacagagttcaaggtagaccaaaccaatgacttaacccataaagacgattgttcccaacgtagatgagaagatttcgaagttctggtgg
L H W L S Q V P S G L V T E L G I S A N K G C I Y S S K A S R P P
159
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
610 620 630 640 650 660 670 680 690 700
* * * * * * * * * *
601 aattcaaaaggcctacttggatcaattcactaaggatttcaccactttcttgagaatccactccgaagaattgatcagtagaggtagaatgttgttgacc 700
ttaagttttccggatgaacctagttaagtgattcctaaagtggtgaaagaactcttaggtgaggcttcttaactagtcatctccatcttacaacaactgg
I Q K A Y L D Q F T K D F T T F L R I H S E E L I S R G R M L L T
192
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
710 720 730 740 750 760 770 780 790 800
* * * * * * * * * *
701 tggatctgcaaagaagatgaatttgaaaacccaaactccatcgatttgttggaaatgtccatcaacgatttggttatcgaaggtcacttagaagaagaaa 800
acctagacgtttcttctacttaaacttttgggtttgaggtagctaaacaacctttacaggtagttgctaaaccaatagcttccagtgaatcttcttcttt
W I C K E D E F E N P N S I D L L E M S I N D L V I E G H L E E E K
225
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
810 820 830 840 850 860 870 880 890 900
* * * * * * * * * *
801 agttggactctttcaacgttccaatctatgctccatctaccgaagaagttaagtgcatcgttgaagaagaaggttccttcgaaatcttgtacttggaaac 900
tcaacctgagaaagttgcaaggttagatacgaggtagatggcttcttcaattcacgtagcaacttcttcttccaaggaagctttagaacatgaacctttg
L D S F N V P I Y A P S T E E V K C I V E E E G S F E I L Y L E T
259
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
910 920 930 940 950 960 970 980 990 1000
* * * * * * * * * *
901 ctttaaggttccatacgatgccggtttctctatcgatgatgattatcaaggtagatcccactctccagtttcttgtgatgaacatgctagagctgctcat 1000
gaaattccaaggtatgctacggccaaagagatagctactactaatagttccatctagggtgagaggtcaaagaacactacttgtacgatctcgacgagta
F K V P Y D A G F S I D D D Y Q G R S H S P V S C D E H A R A A H
292
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
1010 1020 1030 1040 1050 1060 1070 1080 1090 1100
* * * * * * * * * *
1001 gttgcttcagttgttagatctattttcgaacctatcgttgcctctcatttcggtgaagctattatgccagatttgtcccatagaattgctaagaatgctg 1100
caacgaagtcaacaatctagataaaagcttggatagcaacggagagtaaagccacttcgataatacggtctaaacagggtatcttaacgattcttacgac
V A S V V R S I F E P I V A S H F G E A I M P D L S H R I A K N A A
325
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
1110 1120 1130 1140 1150 1160 1170 1180 1190 1200
* * * * * * * * * *
1101 ccaaggttttgagatctggtaagggtttttacgactccttgattatttccttggccaaaaagccagaaaagtccgatgtttctgcttggtcacatccaca 1200
ggttccaaaactctagaccattcccaaaaatgctgaggaactaataaaggaaccggtttttcggtcttttcaggctacaaagacgaaccagtgtaggtgt
K V L R S G K G F Y D S L I I S L A K K P E K S D V S A W S H P Q
359
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
1210 1220 1230
* * *
1201 attcgaaaagtgatgatactagtagcggccgctgcag 1237
taagcttttcactactatgatcatcgccggcgacgtc
F E K * *
392
>>>>>>>>>>>>>>>>
CaDXMT1_Strep-tag
>>>>>>>>>>>>>>>>>>>>>
RFC10 Suffix
Theoretical molecular weight: 44473,7 Da (ProtParam)
Probable posttranslational modifications:
- acetylation at serine (second amino acid)
- O-GlcNAc modifikation at several positions
- phosphorylation at several positions
(see http://2d.bjmu.edu.cn/show2d/Proteomics%20tools.asp)
 "
"