Team:Paris Bettencourt/Suicide
From 2012.igem.org
(→Bigger Picture) |
(→Objectives) |
||
| Line 135: | Line 135: | ||
==Objectives== | ==Objectives== | ||
| - | + | Bacteria have developed mechanisms to kill other bacteria in order to reduce competition between themselves in their environment [1]. Some strains of ''E. coli'' produce '''lethal proteins''' called colicins which kill sensitive bacteria, including related species of E. coli. Colicinogenic cells contain '''specific immunity''' against their own toxin, and their colicins only effect cells containing specific receptors on their outer membrane surface. | |
| + | |||
| + | We have exploited this naturally occurring system to construct a well characterized toxin-antitoxin system ([https://2012.igem.org/Team:Paris_Bettencourt/Suicide#Design design]): | ||
#Design an antitoxin part from the colicin E2 immunity gene that confers immunity against the Col E2 activity protein | #Design an antitoxin part from the colicin E2 immunity gene that confers immunity against the Col E2 activity protein | ||
Revision as of 23:42, 26 September 2012
|
Aims : Implement a kill-switch that features population-level suicide and complete genome degradation. System : A synthetic toxin-anti-toxin system based on the wild type Colicin E2 operon. Achievements : We showed that Colicin E2 cells induce cell death in sensitive populations, and that these sensitive populations can be protected by providing them with our engineered immunity protein.
Part K914001 is well characterized and provides immunity to sensitive cells against the Colicin E2 activity protein, but is leaky. Part K914002 is promoterless and allows users to easily plug in the appropriate promoter for their desired purpose.
|
Contents |
Overview
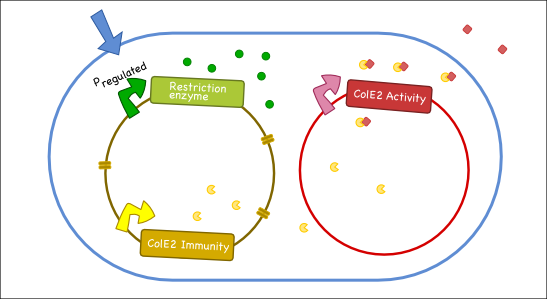
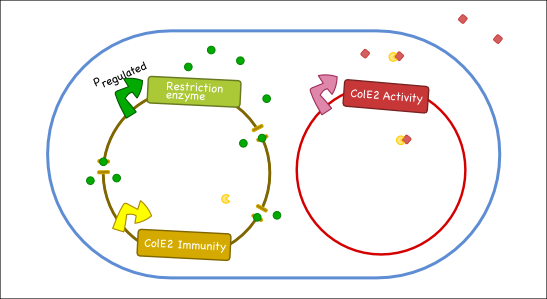
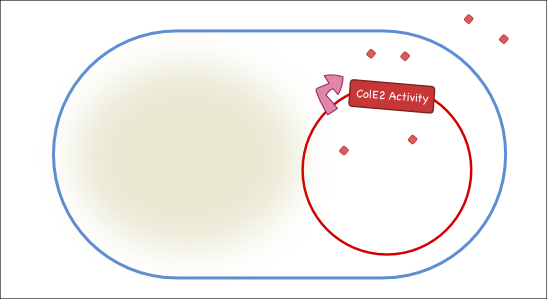
Our goal is to engineer a synthetic toxin-anti-toxin system from the wild type Colicin E2 (Col E2) operon. This synthetic toxin-anti-toxin system is species specific, allows for population-level suicide, complete genome degradation, and will function on a tunable delay. The Col E2 toxin, called the activity protein, is a DNase, meaning that it cleaves DNA, which targets related species of E.coli. The Col E2 anti-toxin, called the immunity protein, binds the Col E2 activity protein with high affinity preventing the activity protein from acting on its own producing bacteria. Our idea is to clone the activity protein and the immunity protein on two different plasmids, called toxin and anti-toxin plasmids, so we can switch on the suicide mechanism by degrading the anti-toxin plasmid with the restriction enzyme system. To test our system, we cloned the immunity protein under the Lac promoter inside a medium copy plasmid. We tested our system by showing that this inducible system protects the cells from the Col E2 activity protein. The next step will be to place the activity protein under a constitutive promoter inside a low copy plasmid; work which is ongoing.
Background
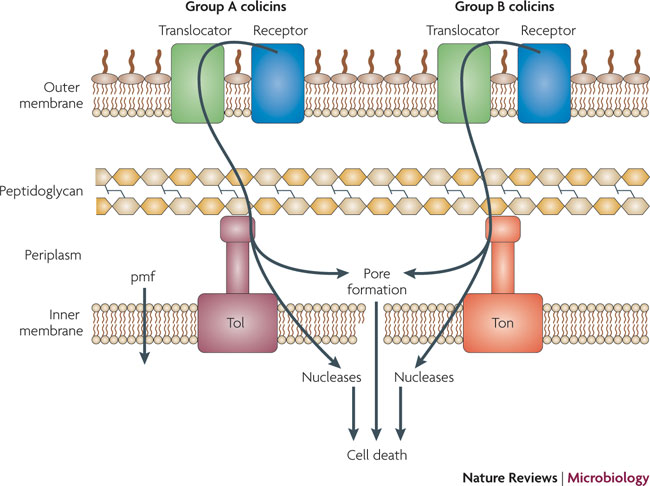
What are Colicins? Bacteria have developed mechanisms to kill other bacteria in order to reduce competition between themselves in the environment [1]. Some strains of E. coli produce lethal proteins called colicins which kill sensitive bacteria, including related species of E. coli. Colicinogenic cells contain specific immunity against their own toxin, and their colicins only effect cells containing specific receptors on their outer membrane surface. Colicins are therefore classified by the receptor to which they bind to, for example, colicins E1-E9 bind to the outer membrane (OM) protein BtuB which mediates the entry of nucleosides, siderphores, and vitamin B12 in the cell. Colicins E1-E9 are also Group A colicins, meaning that they are translocated through the cell envelope by the Tol machinery, whereas Group B colicins are translocated through the cell envelope by the TonB machinery. Colicins have different modes of action, including enzymatic activity such as DNase (DNA cleaving) activity, and pore-forming activity. Colicins have three domains, the N-terminal domain functions in translocation through the membrane, the central domain is involved in binding to the OM receptor, and the C-terminal domain contains active (lethal) protein region [1]. We have taken advantage of the modularity of the colicin domains, generating a synthetic import domain that will allow for new and exciting forms of communication in E.coli.
For our project we have selected Colicin E2, which has enzymatic DNase activity. Colicin E2 is ideal because it not only kills sensitive cells on a population level, but also destroys genomic material, preventing the spread of genetically modified parts via horizontal gene transfer.
In natural systems colicin operons are regulated under the SOS promoter (Stress response). Group A colicins contain type I plasmids. These plasmids are generally 6-10 kb, found in about 20 copies per cell, and can be amplified and mobilized in the presence of a conjugative plasmid. The first gene of the colicin operon is the activity protein, named Colicin X activity (i.e. cea for Colicin E2) . This is followed by the immunity protein, named Colicin X immunity (cei), which, like the activity protein, is regulated by the LexA promoter, but also has its own constitutive promoter. This separate promoter is located with in the coding sequence of the cea and allows for constant overproduction of the cei, thereby protecting the producing cell. The last gene codes for the lysis protein, named colicin X lysis protein (cel), which causes the release of colicin into medium and is responsible for the cell death of the producer. The SOS promoter can be induced by UV light, chemicals, and stress conditions[1].
We would like to clone the cea and cei of Colicin E2 onto two separate plasmids, creating toxin and antitoxin plasmids.
Bigger Picture
Our antitoxin plasmid can be combined with the restriction enzyme system to generate a toxin-antitoxin system that works on a tunable delay depending on the induction of the restriction enzyme. Once the degradation of the antitoxin plasmid containing the immunity protein is initiated, the cell will continue to produce the activity protein, which will digest the cells own genetic material as well as its neighbors and extracellular DNA via its DNase activity.
Objectives
Bacteria have developed mechanisms to kill other bacteria in order to reduce competition between themselves in their environment [1]. Some strains of E. coli produce lethal proteins called colicins which kill sensitive bacteria, including related species of E. coli. Colicinogenic cells contain specific immunity against their own toxin, and their colicins only effect cells containing specific receptors on their outer membrane surface.
We have exploited this naturally occurring system to construct a well characterized toxin-antitoxin system (design):
- Design an antitoxin part from the colicin E2 immunity gene that confers immunity against the Col E2 activity protein
- Measure viability of immune and vulnerable cells in the presence of WT Col E2 cells
- Design a Col E2 activity protein part under the control of a constitutive promoter and establish in immune strains to generate synthetic Col E2 cells
- Measure viability of immune and vulnerable cells in the presence of synthetic Col E2 cells
- Combine our system with the Restriction Enzyme System, measure viability
Design
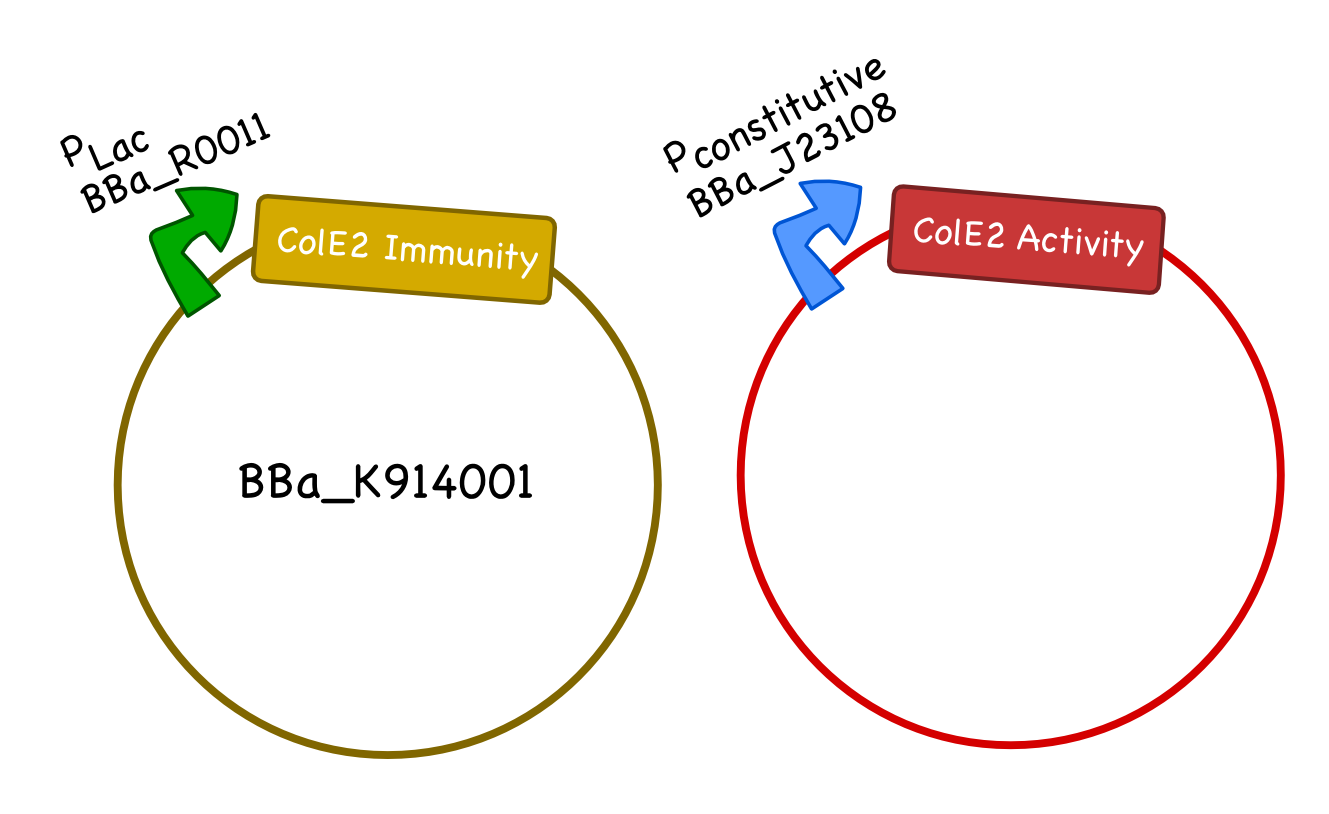
For our system we will generate two plasmids, one carrying the Col E2 activity protein and one carrying the Col E2 immunity protein. As the immunity protein is normally present in excess in natural colicinogenic cells, we chose a 10-12 copy vector pSB3C5 to carry the immunity protein, and a lower ~5 copy vector pSB4K5 to carry the toxin protein. We used pLac to drive expression of the immunity protein, however, different inducible promoters should be used depending on the overall design and application of the safety circuit. We chose the constitutive promoter BBa_J23108 from the Anderson Promoter Collection, which has a relatively moderate expression level, to drive the expression of the activity protein so that it would not overwhelm the cells until after the desired delay. We ultimately plan to integrate the activity protein cassette into the genome of our bacteria.
We do not require the colicin lysis protein for our system, and while the activity protein may degrade extracellular DNA released from our genetically modified bacteria, it is preferable that the cells do not lyse and the genomic material is digested within the cells. A small percentage of cells will die naturally and release the activity protein, which will have a lethal action on any cells that escape our system via mutation of the activity protein gene.
Experiments and results
Colicin E2 Expression Kills Sensitive Cells
We performed a basic assay to characterize the toxicity of the Col E2 cells. Here we expect zones of inhibition (ZOI, clear rings around the colicin cells) when Col E2 is spotted on vulnerable cells.
Experimental setup
Cell Types
- MG1655: These cells approximate wild type E.coli cells as they have undergone minimal genetic manipulation as compared to other laboratory strains. Genotype: F- λ- ilvG- rfb-50 rph-1
- MG1655.RFP: MG1655 cells transformed with constitutive RFP (from Anderson promoter library, [http://partsregistry.org/wiki/index.php/Part:BBa_J23102 BBa_J23102])
- Col E2: Wild Type Colicin E2 cells, containing pColE2-P9 plasmid [2]
Protocol
- Plate 10 uL of 0.1 OD lawn cells from liquid culture on plain LB plates
- Spot 5 uL of saturated Colicin E2 cells from liquid culture on plates
- Incubate plates overnight
Results
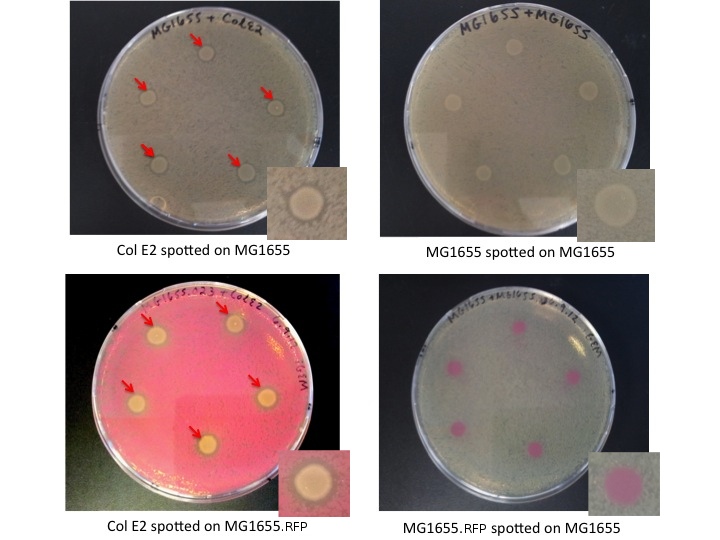
Col E2 cells produce ZOI in sensitive cells, as indicated bellow by red arrows. MG1655 and MG1655.RFP, cells which are not colicinogenic, do not produce ZOI when spotted on a lawn of sensitive cells.
Interpretation
The Col E2 cells secrete the lethal activity protein, which diffuses out from the Col E2 colonies spotted on the LB plates. This generates a region where vulnerable cells can not grow, visible as an empty ring (ZOI) around the Col E2 colonies. It is important to note that Col E2 cells do not need to be induced to produce sufficient activity protein to generate the ZOI. These results confirm the toxicity of Col E2 cells against MG1655 and MG1655.RFP, related species of E.coli. We also assayed Top10 cells, NEB turbo cells, MAGE cells, and MG1655 ZI.RFP cells, for more details look [http://openwetware.org/wiki/IGEM:Paris_Bettencourt_2012/Notebooks/suicide_group/day_by_day//2012/08/07 here]
Expression of Immunity Protects Sensitive Cells
To test the function of our anti-toxin plasmid, we performed Colicin E2 toxicity assays on cells transformed with our plasmid. Here we expect zones of inhibition (ZOI) when Col E2 is spotted on vulnerable cells, and the absence of ZOI in the transformed immune cells.
Experimental setup
Cell Types
- MG1655: These cells approximate wild type E.coli cells as they have undergone minimal genetic manipulation as compared to other laboratory strains. Genotype: F- λ- ilvG- rfb-50 rph-1
- MG1655.RFP: MG1655 cells transformed with constitutive RFP (from Anderson promoter library, BBa_J61002)
- Col E2: Wild Type Colicin E2 cells, containing pColE2-P9 plasmid [2]
- NEB.Imm: NEB turbo cells transformed with the anti-toxin plasmid ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K914001 BBa_K914001] inside [http://partsregistry.org/Part:pSB3C5 pSB3C5]). This strain is commonly used in the lab as it grows rapidly (visible colonies on agar, ~6.5 hours; shaking liquid culture OD 600 = 2.0, ~4 hours). This strain is also T1 phage resistant and importantly it expresses the Lac repressor (lacR), which allows us to induce the production of the immunity protein the pLac promoter with IPTG. Genotype: F´ proA+B+ lacIq ∆ lacZ M15/ fhuA2 ∆(lac-proAB) glnV gal R(zgb-210::Tn10)TetS endA1 thi-1 ∆(hsdS-mcrB)5
References:New England Biolabs, product catalogue number C2984H
- Z1.Imm: MG1655 Z1 cells transformed with the anti-toxin plasmid ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K914001 BBa_K914001] inside [http://partsregistry.org/Part:pSB3C5 pSB3C5]). MG1655 Z1 contain the Z1 cassette (lacR tetR SpR), which allows us to induce the production of the immunity protein under control of the pLac promoter with IPTG.
Protocol
- Grow cells in liquid culture containing the appropriate antibiotic
- Add IPTG (0.1mM) to appropriate liquid cultures after 2 hours (OD ~0.2)
- Wash cells containing antibiotics/IPTG and re-suspend in plain LB
- Plate 10 uL of 0.1 OD lawn cells from liquid culture on plain LB plates or IPTG plates
- Spot 5 uL of saturated Colicin E2 cells from liquid culture on plates
- Incubate plates overnight
Results
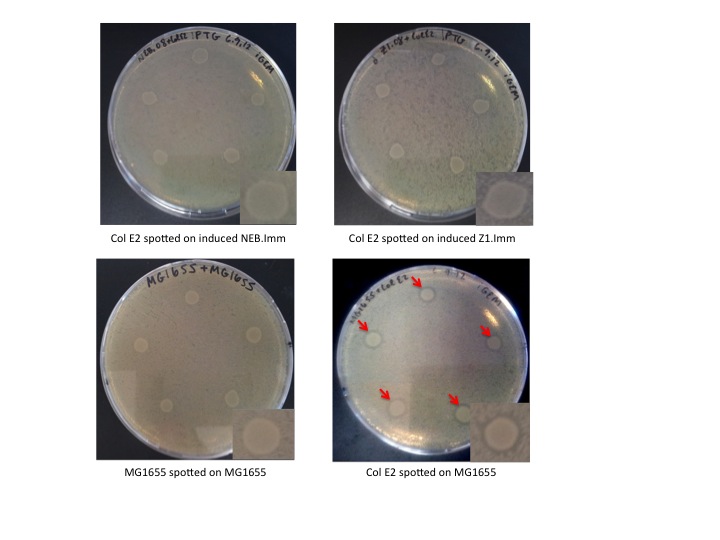
Col E2 cells do not generate a ZOI in cells containing the anti-toxin plasmid, in both induced and un-induced cells. However, Col E2 cells do produce a ZOI in vulnerable MG1655 cells as indicated by red arrows below.
Interpretation
These results indicate that our anti-toxin plasmid indeed confers immunity to sensitive cells, however, immunity is not dependent on IPTG induction. This is not completely surprising given that the immunity protein has an incredibly high affinity for the activity protein and that pLac is known to be leaky. Even a small concentration of immunity protein could protect un-induced cells against the DNase activity of the colicin activity protein.
Quantitative Characterization of the Anti-toxin Plasmid
We characterized the plasmid pLac + ColE2 immunity gene ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K914001 BBa_K914001] inside [http://partsregistry.org/Part:pSB3C5 pSB3C5]) by testing the response of the cells when we mix them with colicin producing cells in various concentration with also various concentration of IPTG as the inducer of pLac. We counted the number of surviving colonies to see the effectiveness of the immunity protein in protecting the cells from the colicin activity protein.
Experimental Setup
Cell types
- Col E2: Wild Type Colicin E2 cells, containing pColE2-P9 plasmid [2]
- NEB.CmR: NEB turbo cells transformed with [http://partsregistry.org/Part:pSB3C5 pSB3C5]).
- NEB.Imm: NEB turbo cells transformed with the anti-toxin plasmid ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K914001 BBa_K914001] inside [http://partsregistry.org/Part:pSB3C5 pSB3C5]).
Protocol
- Grow overnight cultures of NEB.CmR, NEB.Imm each with antibiotic (Cm) and Col E2.
- Centrifuge and resuspend in LB.
- Prepare 5 tubes of 10ml LB and put 40uL of NEB.Imm cells into 4 tubes and NEB.CmR cells in 1 tube.
- Incubate 37C until the OD reaches 0.2.
- Put IPTG with different concentration in 3 NEB.Imm tubes (0.1mM, 0.05mM, 0.025mM).
- Incubate again until the OD reaches 1.0.
- Take each 0.5ml and mix with 0.5ml Colicin cells (saturated, saturated diluted 1000x, plain LB with corresponding amount of IPTG) in small 2ml eppendorf tubes.
- Incubate for 30 minutes.
- Dilute 10000x (100x then 100x) in MgSO4 e-2M.
- Plate 20uL in Cm to kill the Colicin cells and leave the immune cells.
Note: 1ml of cells of OD 1.0 has 1e9 cells, so we expect if all cells survive we will get 1000 colonies at the end.
Results
The table shows the number of colonies survive in each plate IPTG x Colicin.
| 100uM IPTG | 10uM IPTG | 1uM IPTG | 0 IPTG | pSB3C5 (-) control | |
| 1x Colicin | 94 | 79 | 73 | 42 | 0 |
| 1000x Colicin | 9 | 24 | 26 | 74 | 0 |
| No Colicin (+) control | 36 | 28 | 30 | 18 | 27 |
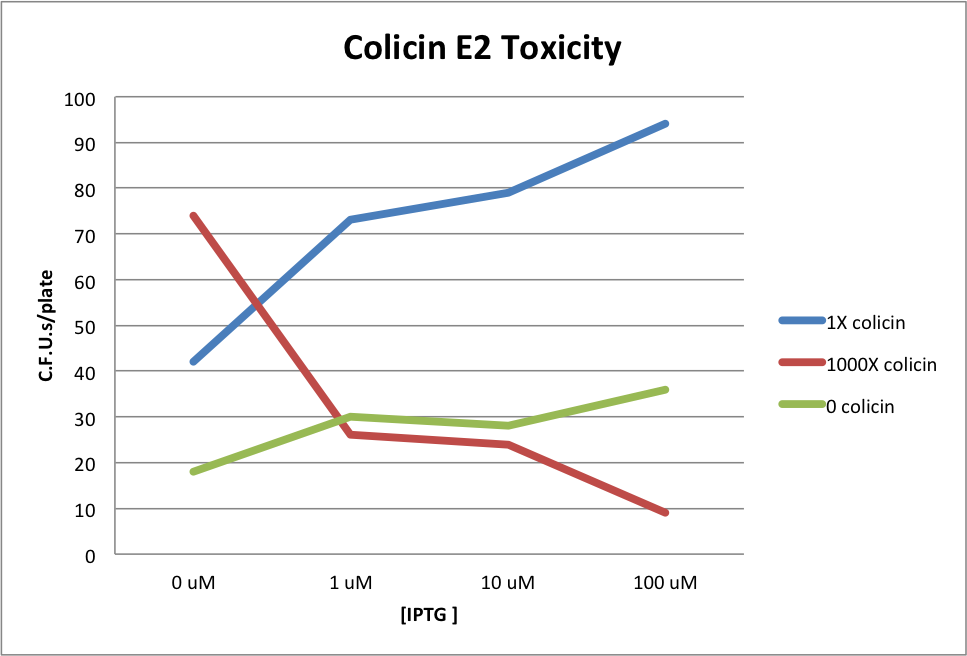
In this experiment we expected to see more survival if we induce the cells with more IPTG and/or mix them with less Colicin. The results showed that the immunity protein can indeed protect the cells from saturated colicin, but the Lac promoter is significantly leaky. Also we noticed that the survival on the positive controls (mixture without Colicin) was somehow less than the one with Colicin. We guessed that the problems are
- The plates have more cells when they have more Colicin: the Colicin cells maybe took the Immunity/Cm-resistance plasmid
- Less cells on the plate without Colicin (and very few cells overall compared to the theoretical calculation): the cells may have died because they loose the Immunity/Cm-resistance plasmid during the incubation without antibiotic (which we did to avoid killing the Colicin cells).
Therefore we still need to improve protocol to provide a more reliable quantitative data.
Perspective
For future experiment, instead of mixing cells with Colicin cells, we could mix them with the supernatant of centrifuged Colicin cells (only the ColE2 protein). Therefore we can put the antibiotic during the whole experiment to avoid plasmid loss. Anyway we have tried to make Colicin supernatant from heat shocked ColE2 cells of OD 1.0 but it failed to kill even the control cells without immunity. We suggest in the future we could give stronger stress to the Colicin to induce more the SOS promoter; such as UV light.
Discussion
Cloning the colicin E2 activity protein, given its lethal nature, is a great challenge for us. We took several approaches. We established immune strains expressing excess immunity protein in which we could transform the toxin plasmid. We tried cloning the toxin without a promoter and finally tried various vectors with and with out promoters. Although the immune strains are viable, transformable, and prove immune to wild type colicin E2 cells, we have yet to successfully transform a single toxin plasmid.
However, developing and characterizing the immune strains was not as straight forward as we imagined. Whether in NEB turbo or MG1655 Z1 cells, the immune strains grew incredibly slowly. Despite being under the control of pLac, their immunity proved somewhat independent of IPTG, as both un-induced and induced cells prove immune. In addition, we had some strange toxicity assay results, partially resolved by the explanation of colicin E2 contamination.
To see more details on the toxicity assays, click [http://openwetware.org/wiki/IGEM:Paris_Bettencourt_2012/Notebooks/suicide_group/day_by_day//2012/09/08 here]
We will continue on our quest to clone the colicin E2 activity protein, and hope that future teams can learn from our experience.
Related Parts and Links
- [http://partsregistry.org/Part:BBa_K131000 BBa_K131000] Colicin E2 operon, designed by Kevin McLeod, group: iGEM08_Calgary_Wetware (2008-07-22)
- [http://partsregistry.org/Part:BBa_R0011 BBa_R0011] Promoter (lacI regulated, lambda pL hybrid), designed by Neelaksh Varshney, Grace Kenney, Daniel Shen, Samantha Sutton, group: Registry (2003-01-31)
- [http://partsregistry.org/Part:BBa_R0011:Experience/iGEM10_Kyoto BBa_R0011:Experience/iGEM10 Kyoto] pLac model
- [http://partsregistry.org/pSB4K5 pSB4K5 ] Low copy plasmid containing kanamycin resistance
- [http://partsregistry.org/pSB3C5 pSB3C5 ] Low-medium copy plasmid containing chloramphenicol resistance
- [http://partsregistry.org/wiki/index.php/Part:BBa_J23108 BBa_J23108] Anderson Constitutive Promoter with relative strength 0.51 (RFP reporter)
- [http://partsregistry.org/wiki/index.php/Part:BBa_J23102 BBa_J23102] Anderson Constitutive Promoter with relative strength 0.86 (RFP reporter)
References
- Cascales E, et al. (2007) Colicin biology. Microbiol Mol Biol Rev 71:158–229.
- Majeed G, Gillor O, Kerr B, Riley MA. (2011) Competitive interactions in Escherichia coli populations: the role of bacteriocins. The ISME Journal 5, 71-81.
- Pugsley AP. (1985) Escherichia coli K12 strains for use in the identification and characterication of colicins. J Gen Microbiol 131: 369-376.
- Kleanthous C. (2010) Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat Microbiol Rev 8:843-848.
Appendix
ColicinE2 Plasmid Map
 "
"


 Overview
Overview Delay system
Delay system Semantic containment
Semantic containment Restriction enzyme system
Restriction enzyme system MAGE
MAGE Encapsulation
Encapsulation Synthetic import domain
Synthetic import domain Safety Questions
Safety Questions Safety Assessment
Safety Assessment