Team:TU Munich/Project/Genome Integration
From 2012.igem.org
(→Results) |
(→Background and principles) |
||
| (17 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
---- | ---- | ||
[[File:Martin_einzel_TUM12.jpg|200px|thumb||Responsible: Martin Schappert]] | [[File:Martin_einzel_TUM12.jpg|200px|thumb||Responsible: Martin Schappert]] | ||
| - | As we | + | As we cannot obey the letter of the German purity law (there is a zero tolerance policy concerning transgenic ingredients), we try our best to meet the spirit. Thus, it is '''unacceptable for us to work with antibiotics''' to keep up the selective environment. Since we cannot work with '''auxotrophies in beer''' either, we have to make sure the yeasts do not lose the BioBricks. The most promising way to accomplish a long lasting presence of our constructs is to achieve '''genome integration'''. |
| - | By means of genome integration we | + | By means of genome integration we do not have to induce a selective stress in the brewing process and yet can be sure that the yeasts express our constructs properly - under [[Team:TU_Munich/Project/Constitutive_Promoter|constitutive promoters]] like '''TEF1 or TEF2''' as well as [[Team:TU_Munich/Project/Ethanol_Inducible_Promoter|ethanol-inducible promoter]] or the [[Team:TU_Munich/Project/Light_Switchable_Promoter|light-switchable promoter]]. |
== Background and principles == | == Background and principles == | ||
---- | ---- | ||
<div> | <div> | ||
| - | To accomplish genome integration we could resort to a preexisting | + | To accomplish genome integration we could resort to a preexisting BioBrick [http://partsregistry.org/Part:BBa_K300001]. |
<div style="float:right"> | <div style="float:right"> | ||
'''BBa K300001 (Pavia '10)''' | '''BBa K300001 (Pavia '10)''' | ||
| Line 22: | Line 22: | ||
* "The part of interest, contained in the yeast integrative vector, will be inserted in the genomic region comprised between the two target sequences by homologous recombination, thus disrupting the original genomic DNA between them."[http://partsregistry.org/wiki/index.php/Part:BBa_K300986] | * "The part of interest, contained in the yeast integrative vector, will be inserted in the genomic region comprised between the two target sequences by homologous recombination, thus disrupting the original genomic DNA between them."[http://partsregistry.org/wiki/index.php/Part:BBa_K300986] | ||
* The recombinant region includes the divergent GAL1/GAL10 promoter and several base pairs of the GAL1 and GAL10 ORFs. | * The recombinant region includes the divergent GAL1/GAL10 promoter and several base pairs of the GAL1 and GAL10 ORFs. | ||
| - | * With SbfI linearized plasmids show a 10- to 50-fold higher integration efficiency than | + | * With SbfI linearized plasmids show a 10- to 50-fold higher integration efficiency than non-linearized ones [https://www.urmc.rochester.edu/labs/Sherman-Lab/publications/pdfs/Saccharomyces-Cerevisiae-Yeast-Intro.pdf] |
* As the plasmid itself contains no yeast ORI, colonies proliferating on G418 plates must have integrated the resistance gene. | * As the plasmid itself contains no yeast ORI, colonies proliferating on G418 plates must have integrated the resistance gene. | ||
</div> | </div> | ||
| Line 28: | Line 28: | ||
== Idea == | == Idea == | ||
---- | ---- | ||
| - | |||
| - | |||
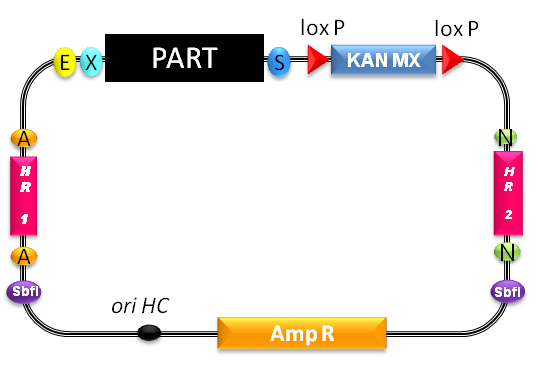
| - | < | + | For proof of principle, we want to insert a functional mOrange cassette (BBa K300007), cut the integrative vector with SbfI in order to linearize it and transfect it into yeasts. After we've shown the general functionality, we'll work on and integrate one gene after another until all of our BioBricks are integrated. In order to do that, we have to engineer new homologous regions to enable multiple transgenic integrations. |
| + | |||
| + | <br> | ||
| + | [[File:Integrational_vector_method_TUM12.jpg|right|406px]] | ||
| + | |||
| + | |||
| + | To show, that the stable integration worked, we picked 36 clones from the plates with yeasts transformed with the linearized integration vector plasmid (insert: BBa K300007[http://partsregistry.org/Part:BBa_K300007]). | ||
| + | These 36 clones were put into non-selective YPD medium and passaged 4 times. After 48 hours, the last culture was used to inoculate an overnight culture. The OD of all candidates was measured. | ||
| + | We had 5 negatives, probably due to mistakes in pipetting. The rest of the cultures were diluted to OD 0.4 and 100 ml of it were used to plate on selective (meaning G418+) petri dishes. | ||
== Results == | == Results == | ||
---- | ---- | ||
| - | |||
| - | |||
| - | |||
| - | Of the 31 remaining plates, 21 were empty. The cfu of the positive plates were: | + | Of the 31 remaining plates, 21 were empty. The cfu of the positive plates were: [[File:Genome_integration_cfu_TUM12.png|right|350px]] |
| + | |||
* 3 | * 3 | ||
* 8 | * 8 | ||
| + | * 12 | ||
* 49 | * 49 | ||
* 68 | * 68 | ||
* 282 | * 282 | ||
* >1000 | * >1000 | ||
| + | * >10 000 | ||
* >10 000 | * >10 000 | ||
* >10 000 | * >10 000 | ||
The plan is to PCR at least two of the clones to gain knowledge why there's been such drastic differences in between the different clones regarding the in the integration efficiency and endurance. | The plan is to PCR at least two of the clones to gain knowledge why there's been such drastic differences in between the different clones regarding the in the integration efficiency and endurance. | ||
| - | |||
| - | |||
As of the mOrange cassette (BBa K300007[http://partsregistry.org/Part:BBa_K300007]), we couldn't show any fluorescence which suggests a corrupt insert. | As of the mOrange cassette (BBa K300007[http://partsregistry.org/Part:BBa_K300007]), we couldn't show any fluorescence which suggests a corrupt insert. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Latest revision as of 20:50, 23 October 2012



Contents |
Genome Integration
As we cannot obey the letter of the German purity law (there is a zero tolerance policy concerning transgenic ingredients), we try our best to meet the spirit. Thus, it is unacceptable for us to work with antibiotics to keep up the selective environment. Since we cannot work with auxotrophies in beer either, we have to make sure the yeasts do not lose the BioBricks. The most promising way to accomplish a long lasting presence of our constructs is to achieve genome integration.
By means of genome integration we do not have to induce a selective stress in the brewing process and yet can be sure that the yeasts express our constructs properly - under constitutive promoters like TEF1 or TEF2 as well as ethanol-inducible promoter or the light-switchable promoter.
Background and principles
To accomplish genome integration we could resort to a preexisting BioBrick [http://partsregistry.org/Part:BBa_K300001].
Summary of principle:
- "The part of interest, contained in the yeast integrative vector, will be inserted in the genomic region comprised between the two target sequences by homologous recombination, thus disrupting the original genomic DNA between them."[http://partsregistry.org/wiki/index.php/Part:BBa_K300986]
- The recombinant region includes the divergent GAL1/GAL10 promoter and several base pairs of the GAL1 and GAL10 ORFs.
- With SbfI linearized plasmids show a 10- to 50-fold higher integration efficiency than non-linearized ones [1]
- As the plasmid itself contains no yeast ORI, colonies proliferating on G418 plates must have integrated the resistance gene.
Idea
For proof of principle, we want to insert a functional mOrange cassette (BBa K300007), cut the integrative vector with SbfI in order to linearize it and transfect it into yeasts. After we've shown the general functionality, we'll work on and integrate one gene after another until all of our BioBricks are integrated. In order to do that, we have to engineer new homologous regions to enable multiple transgenic integrations.
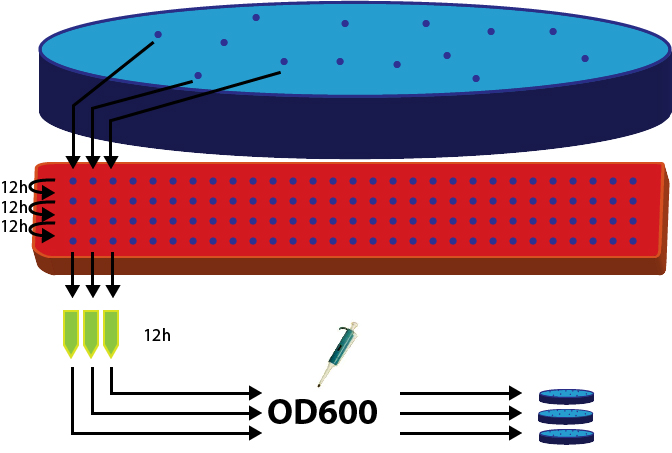
To show, that the stable integration worked, we picked 36 clones from the plates with yeasts transformed with the linearized integration vector plasmid (insert: BBa K300007[http://partsregistry.org/Part:BBa_K300007]).
These 36 clones were put into non-selective YPD medium and passaged 4 times. After 48 hours, the last culture was used to inoculate an overnight culture. The OD of all candidates was measured.
We had 5 negatives, probably due to mistakes in pipetting. The rest of the cultures were diluted to OD 0.4 and 100 ml of it were used to plate on selective (meaning G418+) petri dishes.
Results
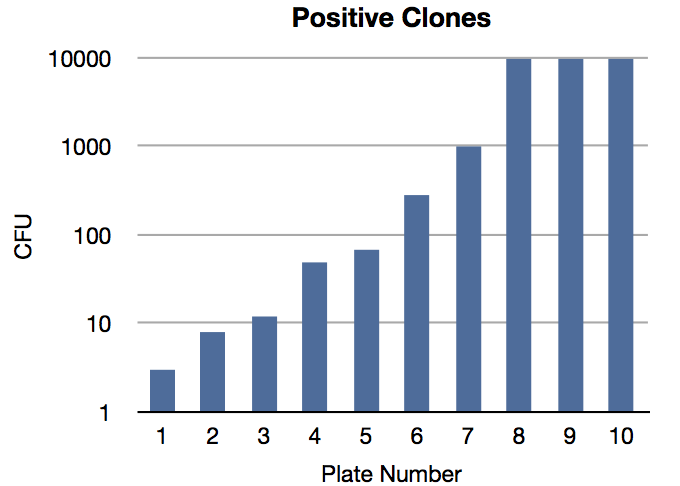
Of the 31 remaining plates, 21 were empty. The cfu of the positive plates were:
- 3
- 8
- 12
- 49
- 68
- 282
- >1000
- >10 000
- >10 000
- >10 000
The plan is to PCR at least two of the clones to gain knowledge why there's been such drastic differences in between the different clones regarding the in the integration efficiency and endurance.
As of the mOrange cassette (BBa K300007[http://partsregistry.org/Part:BBa_K300007]), we couldn't show any fluorescence which suggests a corrupt insert.
 "
"