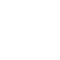Team:Tianjin/Notebook
From 2012.igem.org

July 6th, 2012
- We discuss our all of the possible projects, and finally choose one. And then allocate works to specific member.
- Do some pre-experiment and make some reagants.
- Liquid LB
- 10g tryptone
- 5g yeast extract
- 10g NaCl
- Solid LB
- 10g tryptone
- 5g yeast extract
- 10g NaCl
- 20g agar
- Liquid LB
- Cleaned up the lab and laboratory instruments
July 8th, 2012
- Solutions for extracting plasmid.
- 50mM Glucose / 25mM Tris-Cl / 10mM EDTA,pH=8.0
- 0.2N NaOH / 1% SDS
- 3M KOAc / 2M HOAc
- Experiment technologies training, such as making gel, gel electrophoresis.
July 11th, 2012
- Designed primers and sent orders.
- Optimized the project and allocated work.
- Experiment technologies and knowledge training
- PCR principle and operation
- Gel Extraction
July 13th, 2012
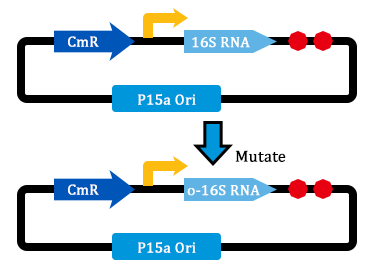
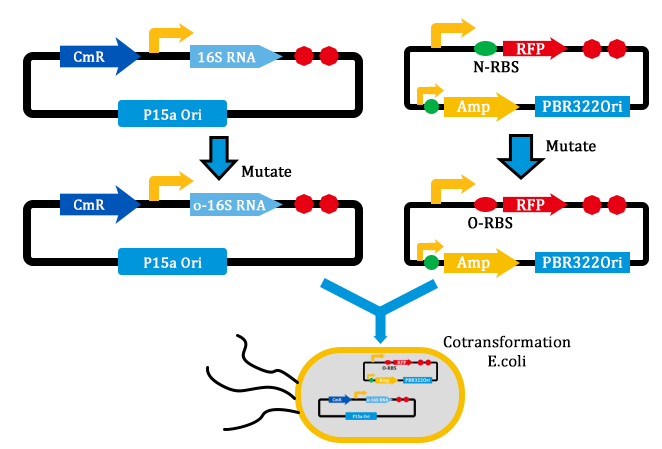
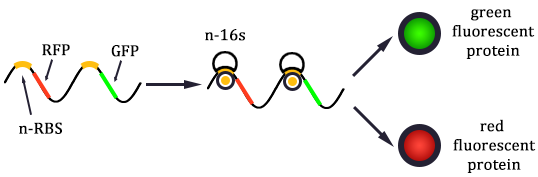
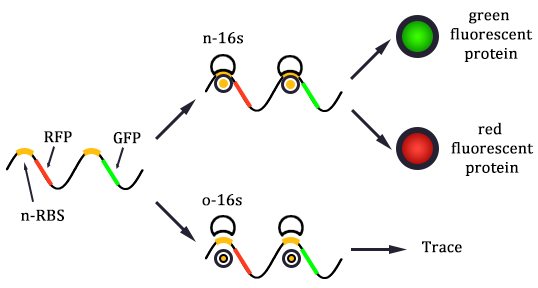
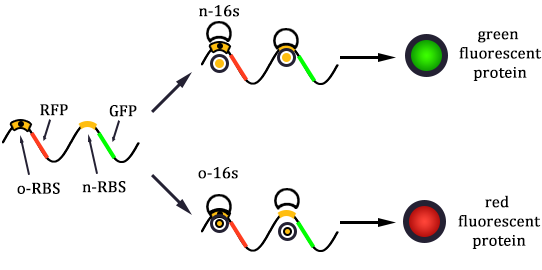
1. Received the oligo and began to mutate 16S rrnB operator.
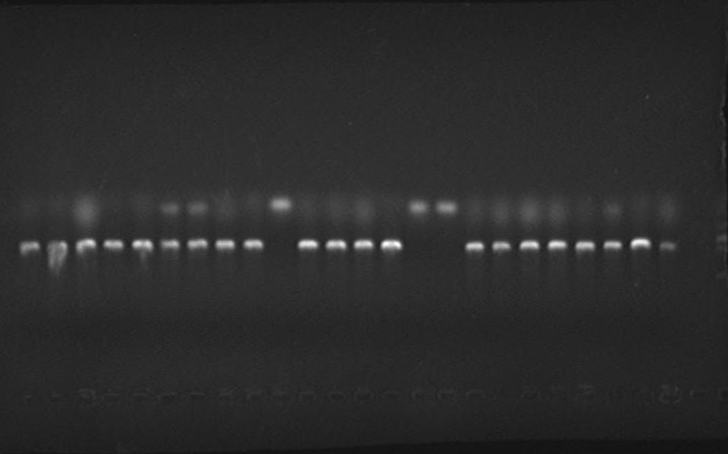
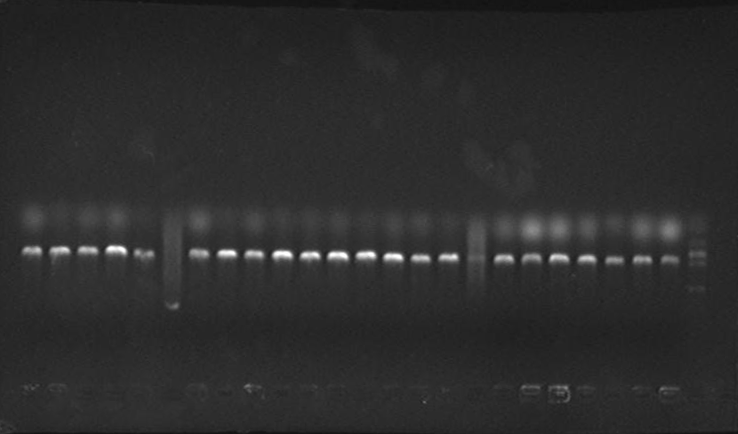
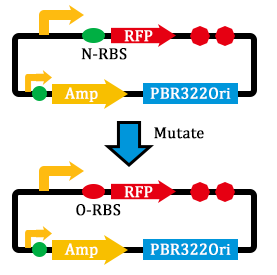
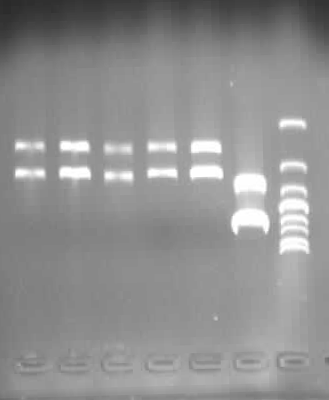
2. Mutated RBS of RFP and checked through gel electrophoresis.
July 14th, 2012
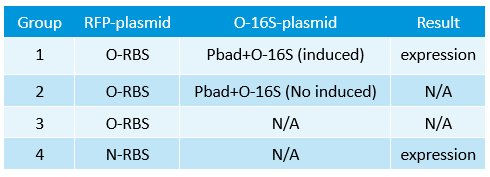
- Transform two plasmids of we got yestday together into E.coli.
- PCR verification of colonies on the LB plates.
- Inculcated the right colonies in Liquid LB.
July 15th, 2012
- Extract plasmid of cells inculcated after one day.
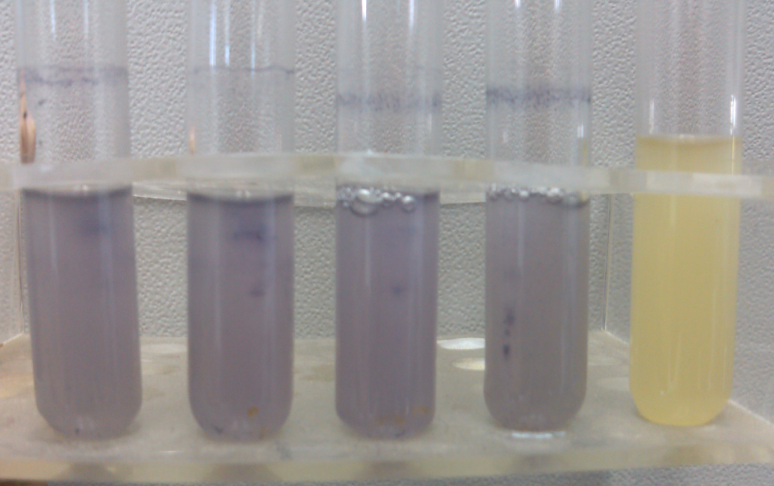
- Enzyme test of the extracted plasmid.
- Inclucated the right cells in Liquid LB and added Ala partly.
July 20th, 2012
Analyse the results.
July 24th, 2012
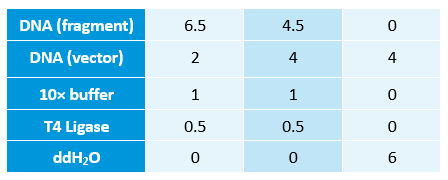
- Linked the GRP gene and O-RBS RFP gene. Gel extract the previous product again, and ligate using T4 with the following gradient.
- In the same method to construct three other systems.
July 30th, 2012
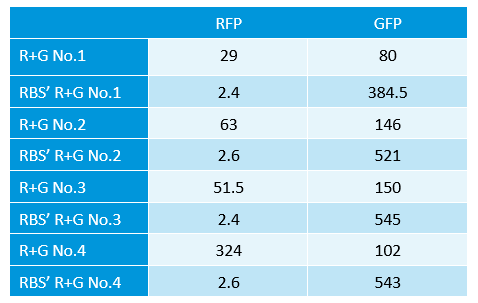
- Learned Fluorescence spectrophotometer.
- measure the strength of GFP and RFP.
Aug. 2nd, 2012
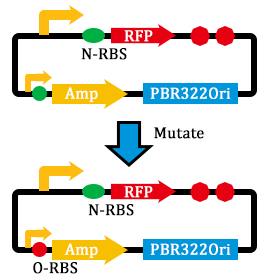
- mutated the RBS of the Amp resistance gene.
- linked genes and transformed them into E. coli.
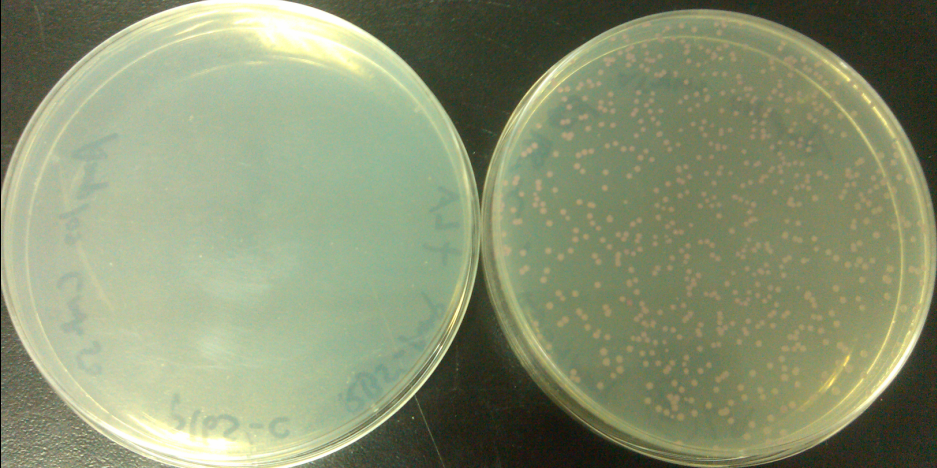
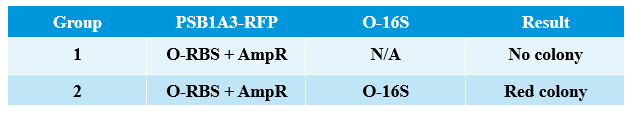
Aug. 4th, 2012
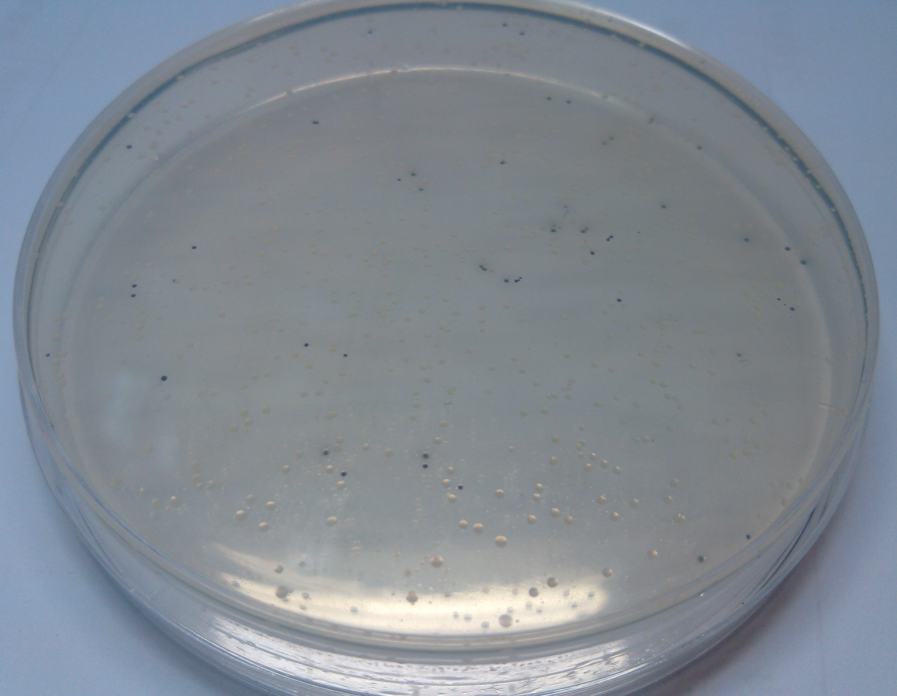
Results of plates
Aug. 8th, 2012
Began to construct the five parts needed in Assembler in Yeast. Part 1 and part 2.
Aug. 11th, 2012
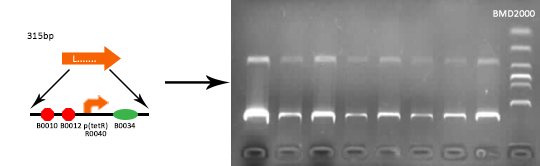
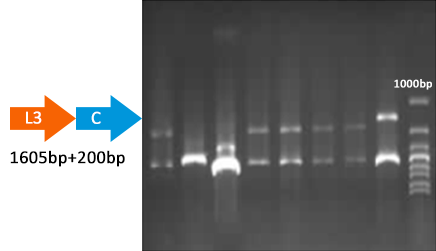
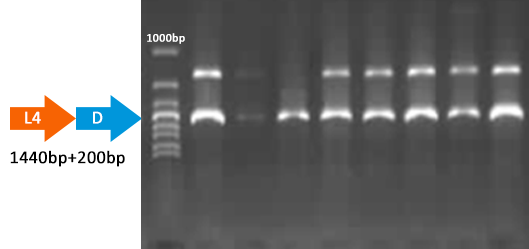
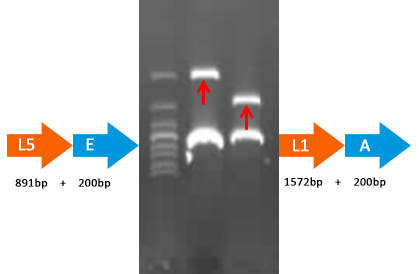
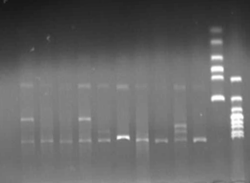
Verified part 1 and 2. Synthesized the two small parts and linked through overlap PCR. Verified through ligase digestion and then gel electrophoresis.
Aug. 12th, 2012
Began to build part 3, 4 and 5. Synthesized the two small parts and linked through overlap PCR.
Aug. 14th, 2012
Verify part 3, 4 and 5. Verified through ligase digestion and then gel electrophoresis.
Aug. 18th, 2012
Transformed all the five parts into Yeast for Assembler in Yeast.
Aug. 20th, 2012
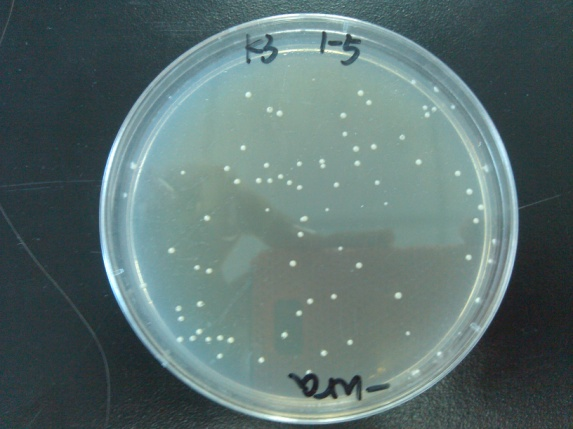
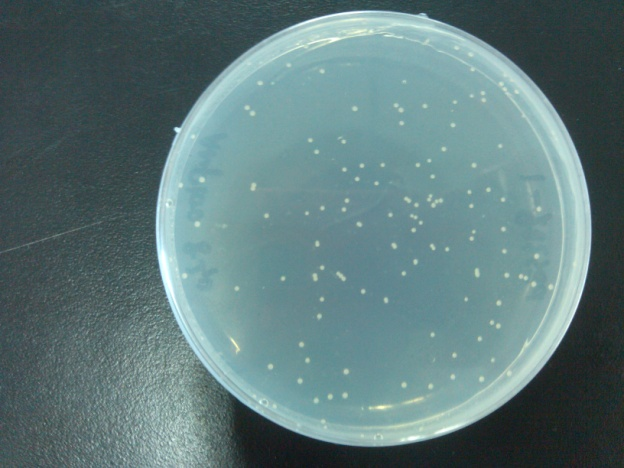
- Results of the transformation.
- Extract plasmids from the Yeast.
Aug. 22nd, 2012
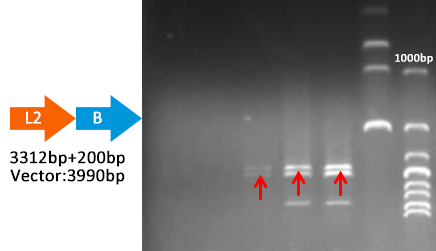
- Transformed the plasmid from the Yeast into cells.
- Extracted plasmid and checked through gel electrophoresis.
- Some other results of Assembler in Yeast.
Aug. 25th, 2012
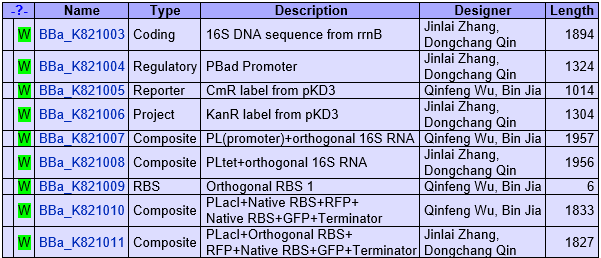
- Began to build biobricks.
- Began to search articles on phages.
Sept. 1st, 2012
Went to Peking University for exchanges.
Sept. 2nd, 2012
- Found out phi X174 and firstly proposed our ideas.
- The second sections of our biobricks.
Sept. 5th, 2012
- Some experiments on phages.
- Optimized our biobricks.
- Designed primers needed for mutating Phi X174 and send orders.
Sept. 11th, 2012
- Began to mutate gene G and E.
- Optimized our biobricks.
Sept. 18th, 2012
Began to sort out our materials for wiki and upload the website.
our team in Hongkong
our gold award
presentation in MIT
 "
"