Team:UTP-Software/Tutorial
From 2012.igem.org
| Home | Team & Attributions | Project | S2MT | Tutorial | Biosinergia | Notebook | Human Practices | Safety | Sponsors |
|---|
|
|
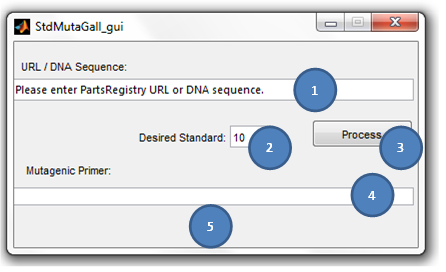
S2MT_championship TutorialS2MT Tutorial - Primer Creation
Some points to take in consideration:
(S2MT) succesfully fixing the incompatibility of the given DNA sequence with the standard 21.
To download the script go to: https://github.com/igemsoftware/UTP_Software_2012 and get all three files. Files descriptions: Note that (S2MT) relies on the Bioinformatics Toolbox of MATLAB (both supplied by iGEM) to make its computations, so you need the toolbox to be able to run the script. You can download this tutorial here: https://static.igem.org/mediawiki/2012/2/29/S2MT_Tutorial.pdf |
 "
"