Team:Tsinghua-A/Wetlab/Part1
From 2012.igem.org
(Difference between revisions)
| (11 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
<link href='http://fonts.googleapis.com/css?family=Oxygen+Mono|Raleway+Dots|Quantico:400' rel='stylesheet' type='text/css'> | <link href='http://fonts.googleapis.com/css?family=Oxygen+Mono|Raleway+Dots|Quantico:400' rel='stylesheet' type='text/css'> | ||
<link rel="stylesheet" type="text/css" href="https://2012.igem.org/Team:Tsinghua-A/css/style-original.css?action=raw&ctype=text/css"/> | <link rel="stylesheet" type="text/css" href="https://2012.igem.org/Team:Tsinghua-A/css/style-original.css?action=raw&ctype=text/css"/> | ||
| - | + | <link rel="stylesheet" type="text/css" href="https://2012.igem.org/Team:Tsinghua-A/css/tt_c.css?action=raw&ctype=text/css"/> | |
<script type="text/javascript" src="https://static.igem.org/mediawiki/2012/c/c8/THU-AIndex.txt"></script> | <script type="text/javascript" src="https://static.igem.org/mediawiki/2012/c/c8/THU-AIndex.txt"></script> | ||
</head> | </head> | ||
| Line 30: | Line 30: | ||
<h2 style="margin-top:135px; ">Construction in Prokaryotic cells</h2> | <h2 style="margin-top:135px; ">Construction in Prokaryotic cells</h2> | ||
<p>In prokaryotic cells, the system consists of three circuits. One induces flip, one flips and the other expresses. According to the properties of the promoter pBAD, we build the first circuit, using arabinose as an inducement to generate the recombinase Cre. After the content of Cre increasing, loxP sites flip, causing the expression of supD tRNA and T7ptag. T7ptag and supD tRNA act as an AND gate which can active T7 promoter by <a href="https://2009.igem.org/Team:PKU_Beijing" style="color:blue;">(iGEM 2009 PKU_Beijing.)</a> Details and truth tables are as follows.</p></br></br> | <p>In prokaryotic cells, the system consists of three circuits. One induces flip, one flips and the other expresses. According to the properties of the promoter pBAD, we build the first circuit, using arabinose as an inducement to generate the recombinase Cre. After the content of Cre increasing, loxP sites flip, causing the expression of supD tRNA and T7ptag. T7ptag and supD tRNA act as an AND gate which can active T7 promoter by <a href="https://2009.igem.org/Team:PKU_Beijing" style="color:blue;">(iGEM 2009 PKU_Beijing.)</a> Details and truth tables are as follows.</p></br></br> | ||
| + | |||
| + | <p style="margin-left:300px; font-size:20px;"><b>Before Reverse</b></p> | ||
| + | |||
<img src="https://static.igem.org/mediawiki/2012/2/20/THU-AWetlab1.png"/> | <img src="https://static.igem.org/mediawiki/2012/2/20/THU-AWetlab1.png"/> | ||
| - | < | + | |
| + | <table border="1" align="center"> | ||
| + | <col width="100"> | ||
| + | <col width="100"> | ||
| + | <col width="100"> | ||
| + | <col width="100"> | ||
| + | <col width="100"> | ||
| + | |||
| + | <tr> | ||
| + | <th> IPTG</th> | ||
| + | <th> aTc</th> | ||
| + | <th> TT-ptag</th> | ||
| + | <th> supD-tRNA </th> | ||
| + | <th> GFP </th> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> 1 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> 0 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 0 </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | |||
| + | </tr> | ||
| + | </table> | ||
| + | |||
</br> | </br> | ||
| + | |||
| + | |||
| + | <p style="margin-left:300px;font-size:20px;"><b>After Reverse</b></p> | ||
| + | |||
<img src="https://static.igem.org/mediawiki/2012/9/97/THU-AWetlab2.png"/> | <img src="https://static.igem.org/mediawiki/2012/9/97/THU-AWetlab2.png"/> | ||
| - | + | ||
</br> | </br> | ||
| - | + | ||
| + | <table border="1" align="center"> | ||
| + | <col width="100"> | ||
| + | <col width="100"> | ||
| + | <col width="100"> | ||
| + | <col width="100"> | ||
| + | <col width="100"> | ||
| + | |||
| + | <tr> | ||
| + | <th> IPTG</th> | ||
| + | <th> aTc</th> | ||
| + | <th> TT-ptag</th> | ||
| + | <th> supD-tRNA </th> | ||
| + | <th> GFP </th> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 0 </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> 1 </td> | ||
| + | <td> 0 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> 0 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | <td> 1 </td> | ||
| + | |||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | </br></br> | ||
| + | |||
| + | </div> | ||
<a href="https://2012.igem.org/Team:Tsinghua-A/Wetlab" style="margin-left:40px;font-size:20px;">Return</a> | <a href="https://2012.igem.org/Team:Tsinghua-A/Wetlab" style="margin-left:40px;font-size:20px;">Return</a> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
Latest revision as of 18:22, 26 September 2012

Tsinghua-A::Wetlab::Prokaryotic cells

Construction in Prokaryotic cells
In prokaryotic cells, the system consists of three circuits. One induces flip, one flips and the other expresses. According to the properties of the promoter pBAD, we build the first circuit, using arabinose as an inducement to generate the recombinase Cre. After the content of Cre increasing, loxP sites flip, causing the expression of supD tRNA and T7ptag. T7ptag and supD tRNA act as an AND gate which can active T7 promoter by (iGEM 2009 PKU_Beijing.) Details and truth tables are as follows.
Before Reverse
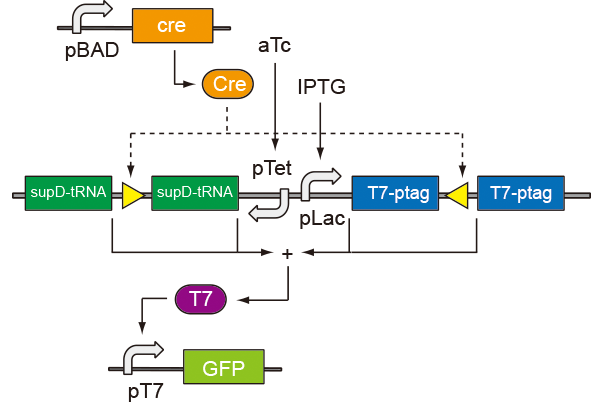
| IPTG | aTc | TT-ptag | supD-tRNA | GFP |
|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 1 | 0 | 0 |
| 0 | 1 | 0 | 1 | 0 |
| 1 | 1 | 1 | 1 | 1 |
After Reverse

| IPTG | aTc | TT-ptag | supD-tRNA | GFP |
|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 1 | 1 | 1 |
| 0 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 |
 "
"