Team:Stanford-Brown/HellCell/pH
From 2012.igem.org
(→pH At a glance Extremophile: Escherichia coli Proteins of interest: serine deaminase (serine catabolism) Consensus: Helpful) |
(→pH At a glance Extremophile: Escherichia coli Proteins of interest: serine deaminase (serine catabolism) Consensus: Helpful) |
||
| Line 20: | Line 20: | ||
What the Hell Cell squad found intriguing, however, is that it has been found that E. coli increases its catabolism of amino acids when exposed to high pHs, most researched of which are tryptophan and serine (Padan et al.). This process creates buffers in the cytoplasm to help counter the effects of the high pH. Since the heightened catabolism of tryptophan would require the insertion of a tryptophan transporter, we decided to choose serine catabolism and isolated the gene for serine deaminase from ''E. coli''. | What the Hell Cell squad found intriguing, however, is that it has been found that E. coli increases its catabolism of amino acids when exposed to high pHs, most researched of which are tryptophan and serine (Padan et al.). This process creates buffers in the cytoplasm to help counter the effects of the high pH. Since the heightened catabolism of tryptophan would require the insertion of a tryptophan transporter, we decided to choose serine catabolism and isolated the gene for serine deaminase from ''E. coli''. | ||
| - | We inserted this gene into the Test Plasmid and | + | We inserted this gene into the Test Plasmid, transformed ''E. coli'' with it, and assay its function. We also tested the effectiveness of two genes from our radiation assays, Dps and recA--see Radiation. |
~stuff about protocol~ | ~stuff about protocol~ | ||
| - | [[File:Base_graph.jpeg]] | + | [[File:Base_graph.jpeg|800px|center]] |
~analysis of data~ | ~analysis of data~ | ||
Sources: Padan, E., Bibi, E., Ito, M., Krulwich, T.A. (2005). Alkaline pH Homeostasis in Bacteria: New Insights. ''Biochim Biophys Acta, 1717''(2): 67-88. | Sources: Padan, E., Bibi, E., Ito, M., Krulwich, T.A. (2005). Alkaline pH Homeostasis in Bacteria: New Insights. ''Biochim Biophys Acta, 1717''(2): 67-88. | ||
Revision as of 00:36, 2 October 2012
pH
At a glance
Extremophile: Escherichia coli
Proteins of interest: serine deaminase (serine catabolism)
Consensus: Helpful
Alkaliphiles like marine bacteria (pH 8.2) and pyloric duct flora (pH ~10 or higher) rely largely on transmembrane transporter proteins to regulate the pH within the cytoplasm and thrive (Padan, Bibi, Ito, and Krulwich 2005). Most notable of these transporters are ATP synthase and cation/proton antiporters. Both of these use some type of energy to power the transport of H+ ions across the membrane: in the former, the energy released in the dephosphorylation of ATP is coupled with the transport; in the latter, an electrochemical gradient is exploited (Padan, et al. 2005). All the genes required for these transmembrane proteins total ~10 kb in length, however, which is difficult to manage in BioBrick form. Additionally, these proteins are optimized for the membranes of alkaliphilic bacteria which have a different lipid composition than E. coli, making their behavior tricky to control in a planned fashion (Padan, et al. 2005).
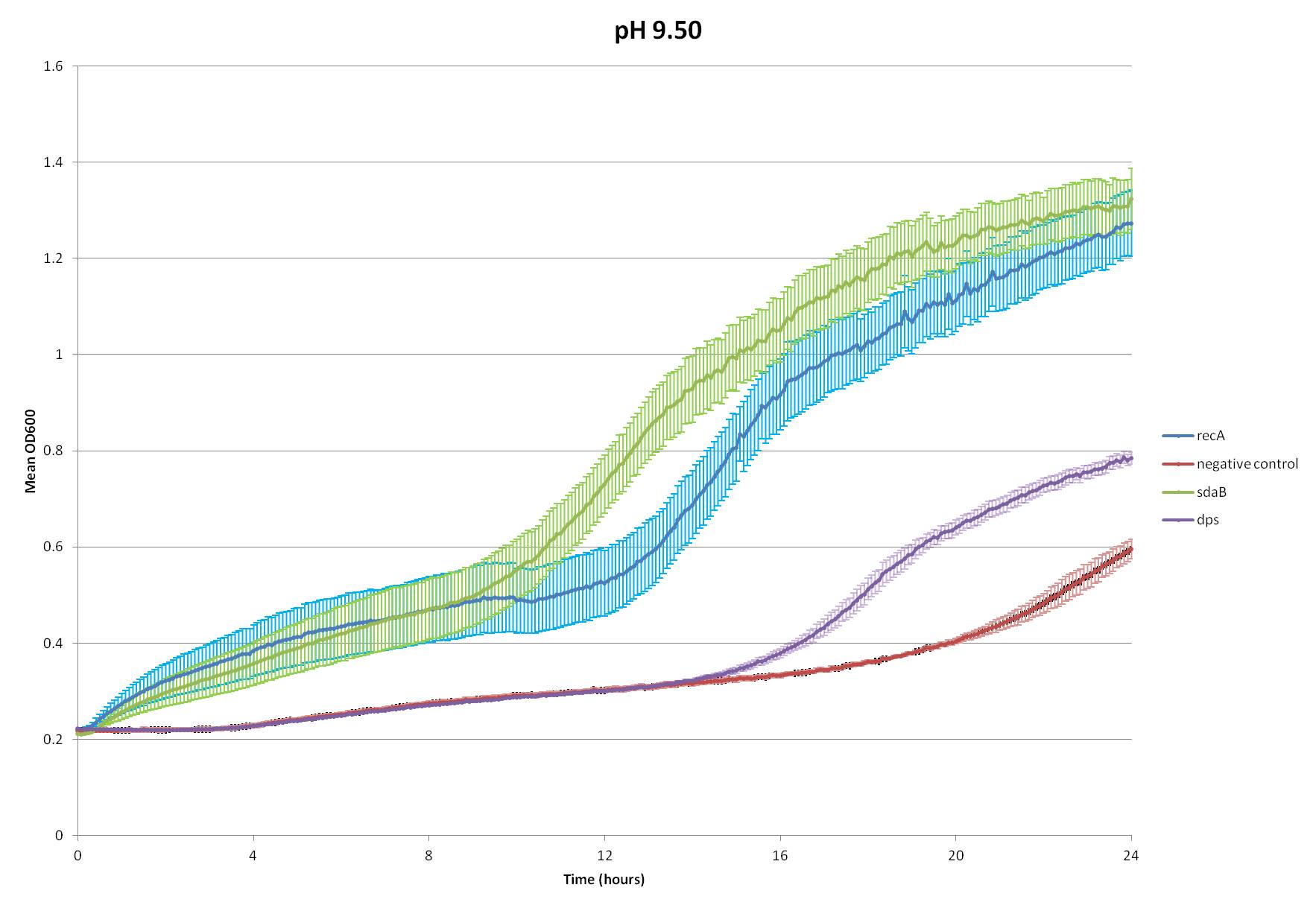
What the Hell Cell squad found intriguing, however, is that it has been found that E. coli increases its catabolism of amino acids when exposed to high pHs, most researched of which are tryptophan and serine (Padan et al.). This process creates buffers in the cytoplasm to help counter the effects of the high pH. Since the heightened catabolism of tryptophan would require the insertion of a tryptophan transporter, we decided to choose serine catabolism and isolated the gene for serine deaminase from E. coli.
We inserted this gene into the Test Plasmid, transformed E. coli with it, and assay its function. We also tested the effectiveness of two genes from our radiation assays, Dps and recA--see Radiation.
~stuff about protocol~
~analysis of data~
Sources: Padan, E., Bibi, E., Ito, M., Krulwich, T.A. (2005). Alkaline pH Homeostasis in Bacteria: New Insights. Biochim Biophys Acta, 1717(2): 67-88.
 "
"