Team:ETH Zurich/Project overview
From 2012.igem.org
m |
|||
| Line 11: | Line 11: | ||
=== ''E.Colipse'' - intelligent sunscreen=== | === ''E.Colipse'' - intelligent sunscreen=== | ||
| - | [[Image:ETH_sunspectrum_lighttypes.png|right|thumb|250px|''Figure'': Spectrum of the sun. Dotted lines indicate the borders between different light types denoted with the labels at the top. 'A', 'B' and 'C' show the regions for UV-A/B/C radiation.]] | + | [[Image:ETH_sunspectrum_lighttypes.png|right|thumb|250px|''Figure 1'': Spectrum of the sun. Dotted lines indicate the borders between different light types denoted with the labels at the top. 'A', 'B' and 'C' show the regions for UV-A/B/C radiation.]] |
Sun UV radiation is unavoidable during warm sunny days. Despite the fact that suns light reaching Earth crust consists of only few percent of UV-A (320 - 400 nm) and UV-B (280 - 320 nm) radiation, it still poses serious risks and life destructive capabilities, e.g. DNA is one of the main targets which are hit by UV radiation, causing DNA breakage, mutations etc. (literature) If unattended, this can lead to sun burns, skin damage or even cancer. Thus, a proper skin protection is usually a good idea in order to avoid unpleasant experience. | Sun UV radiation is unavoidable during warm sunny days. Despite the fact that suns light reaching Earth crust consists of only few percent of UV-A (320 - 400 nm) and UV-B (280 - 320 nm) radiation, it still poses serious risks and life destructive capabilities, e.g. DNA is one of the main targets which are hit by UV radiation, causing DNA breakage, mutations etc. (literature) If unattended, this can lead to sun burns, skin damage or even cancer. Thus, a proper skin protection is usually a good idea in order to avoid unpleasant experience. | ||
| Line 22: | Line 22: | ||
=== Who's your PABA?=== | === Who's your PABA?=== | ||
| - | [[Image:PABA.png|left|thumb|100px|''Figure'': 4-para-aminobenzoic acid, natural occuring UV light absorbing molecule used in sunscreens]] | + | [[Image:PABA.png|left|thumb|100px|''Figure 2'': 4-para-aminobenzoic acid, natural occuring UV light absorbing molecule used in sunscreens]] |
For our sunscreen we chose ''E. coli'' as our primer chassis, with a later idea to transfer a working system to a natural bacterial [https://2012.igem.org/Team:ETH_Zurich/Applications flora of the skin or alginate beads]. As a UV-B light absorbing substance, [https://2012.igem.org/Team:ETH_Zurich/PABA 4-para-aminobenzoic acid] (PABA) was chosen for the following reasons: i) PABA is already used in sunscreens for UV-B protection (literature); ii) PABA is a natural occurring compound as an intermediate in folate synthesis pathway. Furthermore, overproduction of PABA requires overexpression of just two bottleneck enzymes: pabA and pabB, and several attempts were made to overproduce PABA in E.coli by other iGEM teams (literature). Thus we cloned both, [https://2012.igem.org/Team:ETH_Zurich/PABA/Design pabA and pabB, into one operon] under the constitutive promoter aiming for [https://2012.igem.org/Team:ETH_Zurich/PABA/Results increase PABA concentration] in the cells. Furthermore, we are not limited to PABA and other natural light absorbance molecules, such as mycosporine-like amino acids, might be used for a final system (literature: review about biosunscreens. | For our sunscreen we chose ''E. coli'' as our primer chassis, with a later idea to transfer a working system to a natural bacterial [https://2012.igem.org/Team:ETH_Zurich/Applications flora of the skin or alginate beads]. As a UV-B light absorbing substance, [https://2012.igem.org/Team:ETH_Zurich/PABA 4-para-aminobenzoic acid] (PABA) was chosen for the following reasons: i) PABA is already used in sunscreens for UV-B protection (literature); ii) PABA is a natural occurring compound as an intermediate in folate synthesis pathway. Furthermore, overproduction of PABA requires overexpression of just two bottleneck enzymes: pabA and pabB, and several attempts were made to overproduce PABA in E.coli by other iGEM teams (literature). Thus we cloned both, [https://2012.igem.org/Team:ETH_Zurich/PABA/Design pabA and pabB, into one operon] under the constitutive promoter aiming for [https://2012.igem.org/Team:ETH_Zurich/PABA/Results increase PABA concentration] in the cells. Furthermore, we are not limited to PABA and other natural light absorbance molecules, such as mycosporine-like amino acids, might be used for a final system (literature: review about biosunscreens. | ||
| Line 29: | Line 29: | ||
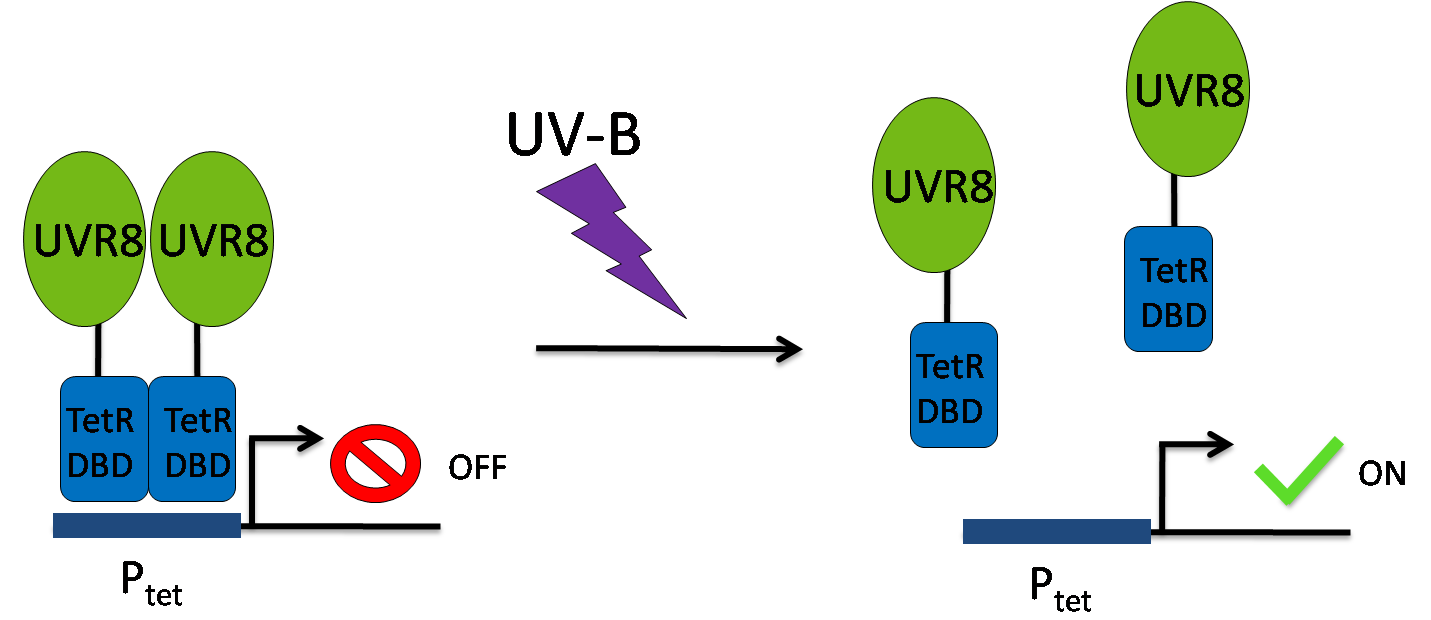
To couple PABA synthesis to UV light we investigated ''E.coli'' response to UV radiation. However, in prokaryotes such response is activated indirectly by sensing DNA damage (literature). Such protection strategy would be erroneous to pursue, because induction of protection is initiated after DNA damage is already happened. Thus we used plant UV-B responsive protein [https://2012.igem.org/Team:ETH_Zurich/UVR8 UVR8] to design a novel UV-B receptor in ''E. coli''. | To couple PABA synthesis to UV light we investigated ''E.coli'' response to UV radiation. However, in prokaryotes such response is activated indirectly by sensing DNA damage (literature). Such protection strategy would be erroneous to pursue, because induction of protection is initiated after DNA damage is already happened. Thus we used plant UV-B responsive protein [https://2012.igem.org/Team:ETH_Zurich/UVR8 UVR8] to design a novel UV-B receptor in ''E. coli''. | ||
| - | In the dark state, [https://2012.igem.org/Team:ETH_Zurich/UVR8 UVR8] forms a dimer, which monomerizes upon UV-B irradiation and induces transcriptional changes necessary for plant UV response (literature). However, UVR8 is not able to bind DNA, thus we fused it with a DNA binding domain of tetracycline repressor protein ([http://partsregistry.org/Part:BBa_K909007 TetR<sub>DBD</sub> ]). TetR<sub>DBD</sub> does not contain dimerization domain required for efficient binding and repression of TetR responsive promoter P<sub>tet</sub>. However, external dimerization would in principle restore TetR<sub>DBD</sub> ability to repress P<sub>tet</sub>. Thus, our chimeric protein [https://2012.igem.org/Team:ETH_Zurich/UVR8/Design UVR8-TetR<sub>DBD</sub>] acts as an light inducible transcription repressor – at dark state, UVR8 dimer brings TetR<sub>DBD</sub> at close proximity to bind P<sub>tet</sub>, whereas UVR8 monomerization upon UV-B light releases UVR8-TetR<sub>DBD</sub> from DNA, thus inducing transcription of the downstream genes (fig.). | + | In the dark state, [https://2012.igem.org/Team:ETH_Zurich/UVR8 UVR8] forms a dimer, which monomerizes upon UV-B irradiation and induces transcriptional changes necessary for plant UV response (literature). However, UVR8 is not able to bind DNA, thus we fused it with a DNA binding domain of tetracycline repressor protein ([http://partsregistry.org/Part:BBa_K909007 TetR<sub>DBD</sub> ]). TetR<sub>DBD</sub> does not contain dimerization domain required for efficient binding and repression of TetR responsive promoter P<sub>tet</sub>. However, external dimerization would in principle restore TetR<sub>DBD</sub> ability to repress P<sub>tet</sub>. Thus, our chimeric protein [https://2012.igem.org/Team:ETH_Zurich/UVR8/Design UVR8-TetR<sub>DBD</sub>] acts as an light inducible transcription repressor – at dark state, UVR8 dimer brings TetR<sub>DBD</sub> at close proximity to bind P<sub>tet</sub>, whereas UVR8 monomerization upon UV-B light releases UVR8-TetR<sub>DBD</sub> from DNA, thus inducing transcription of the downstream genes (fig. 3). |
| - | [[File:UVR8-tetRDBD scheme.png|frameless|500px|center|thumb|''Figure '': UVR8-TetR<sub>DBD</sub> working mechanism: in dark state UVR8-TetR<sub>DBD</sub> is a dimer and represses P<sub>tet</sub>, while under exposure of UV-B light UVR8-TetR<sub>DBD</sub> releases DNA and allows transcription of a downstream genes. ]] | + | [[File:UVR8-tetRDBD scheme.png|frameless|500px|center|thumb|''Figure 3'': UVR8-TetR<sub>DBD</sub> working mechanism: in dark state UVR8-TetR<sub>DBD</sub> is a dimer and represses P<sub>tet</sub>, while under exposure of UV-B light UVR8-TetR<sub>DBD</sub> releases DNA and allows transcription of a downstream genes. ]] |
Our [https://2012.igem.org/Team:ETH_Zurich/Modeling/UVR8 models] suggest that such system would efficiently induce production of PABA at the right time scale, giving broad possibilities for such novel UV-B switch. Also, [https://2012.igem.org/Team:ETH_Zurich/UVR8/Results our data] shows that repression of P<sub>tet</sub> is indeed restored for TetR<sub>DBD</sub>, however, higher inductions of UVR8-TetR<sub>DBD</sub>, required for tighter P<sub>tet</sub> repression, results in the formation of inclusion bodies and cell cytotoxicity. Thus, a second approach of indirect UV sensing was also investigated. | Our [https://2012.igem.org/Team:ETH_Zurich/Modeling/UVR8 models] suggest that such system would efficiently induce production of PABA at the right time scale, giving broad possibilities for such novel UV-B switch. Also, [https://2012.igem.org/Team:ETH_Zurich/UVR8/Results our data] shows that repression of P<sub>tet</sub> is indeed restored for TetR<sub>DBD</sub>, however, higher inductions of UVR8-TetR<sub>DBD</sub>, required for tighter P<sub>tet</sub> repression, results in the formation of inclusion bodies and cell cytotoxicity. Thus, a second approach of indirect UV sensing was also investigated. | ||
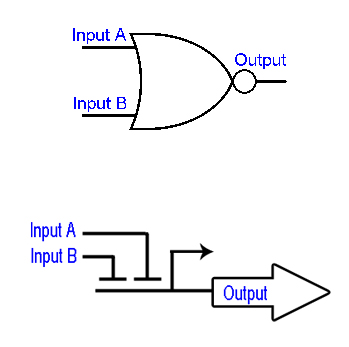
| - | [[Image:NOR_gates.jpg|right|thumb|130px|Figure: NOR gates: Boolean NOR gate representation (upper) and genomic NOR gate - promoter regulated by two repressors (bottom) ]] | + | [[Image:NOR_gates.jpg|right|thumb|130px|''Figure 4'': NOR gates: Boolean NOR gate representation (upper) and genomic NOR gate - promoter regulated by two repressors (bottom) ]] |
=== Indirect UV detection: decoding of sun light from its light spectrum=== | === Indirect UV detection: decoding of sun light from its light spectrum=== | ||
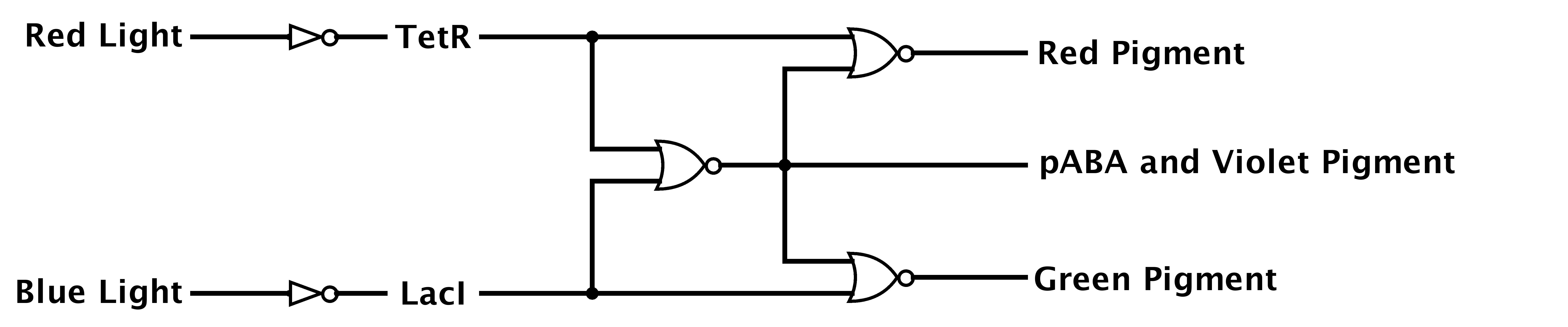
Each light source has its characteristic light emission spectrum e.g. sun contains high red and blue light content compared to artificial light sources, such as tungsten light bulb have just high red and a very small fraction of blue light. Thus, we design [https://2012.igem.org/Team:ETH_Zurich/Decoder a biological circuit] which can distinguish between four different light conditions: i) high red-high blue; ii) high red-low blue; iii) low red-high blue and iv) low red-low blue (fig.). Such circuit consists of [https://2012.igem.org/Team:ETH_Zurich/Decoder/Design three hybrid promoters] each acting as a NOR gate for two different transcriptional repressors i.e. transcription from such promoter is activated just when both repressors are absent. | Each light source has its characteristic light emission spectrum e.g. sun contains high red and blue light content compared to artificial light sources, such as tungsten light bulb have just high red and a very small fraction of blue light. Thus, we design [https://2012.igem.org/Team:ETH_Zurich/Decoder a biological circuit] which can distinguish between four different light conditions: i) high red-high blue; ii) high red-low blue; iii) low red-high blue and iv) low red-low blue (fig.). Such circuit consists of [https://2012.igem.org/Team:ETH_Zurich/Decoder/Design three hybrid promoters] each acting as a NOR gate for two different transcriptional repressors i.e. transcription from such promoter is activated just when both repressors are absent. | ||
| - | We developed a novel [https://2012.igem.org/Team:ETH_Zurich/Modeling/Photoinduction photoinduction model] to select for a suitable light receptors for the system inputs: [https://2012.igem.org/Team:ETH_Zurich/LovTAP LovTAP] (blue light receptor) and [https://2012.igem.org/Team:ETH_Zurich/Cph8 Cph8] (Red light receptor | + | We developed a novel [https://2012.igem.org/Team:ETH_Zurich/Modeling/Photoinduction photoinduction model] to select for a suitable light receptors for the system inputs: [https://2012.igem.org/Team:ETH_Zurich/LovTAP LovTAP] (blue light receptor) and [https://2012.igem.org/Team:ETH_Zurich/Cph8 Cph8] (Red light receptor). Later analysis of the circuit network revealed several [https://2012.igem.org/Team:ETH_Zurich/Modeling/Construct2 kinetic parameters constrains] upon which hybrid promoters were chosen and synthesized. Further testing of the promoters showed that as predicted, they act as a NOR gate, giving high potential of building a first full working light regulated biological decoder circuit in cells. |
| - | [[File:Design.jpg|frameless|700px|center|thumb|Figure : Boolean logic of Decoder. ]] | + | [[File:Design.jpg|frameless|700px|center|thumb|''Figure 5'': Boolean logic of Decoder. ]] |
<!--[[File:DecoderCircuit.png|frameless|500px|center|thumb|''Figure'': Circuit for Decoder. ]]--> | <!--[[File:DecoderCircuit.png|frameless|500px|center|thumb|''Figure'': Circuit for Decoder. ]]--> | ||
{{:Team:ETH_Zurich/Templates/Footer}} | {{:Team:ETH_Zurich/Templates/Footer}} | ||
Revision as of 19:27, 26 October 2012
Contents |
E.Colipse - intelligent sunscreen
Sun UV radiation is unavoidable during warm sunny days. Despite the fact that suns light reaching Earth crust consists of only few percent of UV-A (320 - 400 nm) and UV-B (280 - 320 nm) radiation, it still poses serious risks and life destructive capabilities, e.g. DNA is one of the main targets which are hit by UV radiation, causing DNA breakage, mutations etc. (literature) If unattended, this can lead to sun burns, skin damage or even cancer. Thus, a proper skin protection is usually a good idea in order to avoid unpleasant experience.
However, conventional sunscreens provide relatively passive sun protection, leaving actual protection regulation to ones speculations when and how much of the sunscreen should be applied, which easily can result in inefficient layer of sunscreen and skin damage.
Thus, we tried to develop an intelligent sunscreen, which could adapt its protection factor to the exposed UV radiation and also give a warning signal if protection is insufficient.
Who's your PABA?
For our sunscreen we chose E. coli as our primer chassis, with a later idea to transfer a working system to a natural bacterial flora of the skin or alginate beads. As a UV-B light absorbing substance, 4-para-aminobenzoic acid (PABA) was chosen for the following reasons: i) PABA is already used in sunscreens for UV-B protection (literature); ii) PABA is a natural occurring compound as an intermediate in folate synthesis pathway. Furthermore, overproduction of PABA requires overexpression of just two bottleneck enzymes: pabA and pabB, and several attempts were made to overproduce PABA in E.coli by other iGEM teams (literature). Thus we cloned both, pabA and pabB, into one operon under the constitutive promoter aiming for increase PABA concentration in the cells. Furthermore, we are not limited to PABA and other natural light absorbance molecules, such as mycosporine-like amino acids, might be used for a final system (literature: review about biosunscreens.
Direct UV detection: a novel UV-B light receptor in E.coli
To couple PABA synthesis to UV light we investigated E.coli response to UV radiation. However, in prokaryotes such response is activated indirectly by sensing DNA damage (literature). Such protection strategy would be erroneous to pursue, because induction of protection is initiated after DNA damage is already happened. Thus we used plant UV-B responsive protein UVR8 to design a novel UV-B receptor in E. coli.
In the dark state, UVR8 forms a dimer, which monomerizes upon UV-B irradiation and induces transcriptional changes necessary for plant UV response (literature). However, UVR8 is not able to bind DNA, thus we fused it with a DNA binding domain of tetracycline repressor protein ([http://partsregistry.org/Part:BBa_K909007 TetRDBD ]). TetRDBD does not contain dimerization domain required for efficient binding and repression of TetR responsive promoter Ptet. However, external dimerization would in principle restore TetRDBD ability to repress Ptet. Thus, our chimeric protein UVR8-TetRDBD acts as an light inducible transcription repressor – at dark state, UVR8 dimer brings TetRDBD at close proximity to bind Ptet, whereas UVR8 monomerization upon UV-B light releases UVR8-TetRDBD from DNA, thus inducing transcription of the downstream genes (fig. 3).
Our models suggest that such system would efficiently induce production of PABA at the right time scale, giving broad possibilities for such novel UV-B switch. Also, our data shows that repression of Ptet is indeed restored for TetRDBD, however, higher inductions of UVR8-TetRDBD, required for tighter Ptet repression, results in the formation of inclusion bodies and cell cytotoxicity. Thus, a second approach of indirect UV sensing was also investigated.
Indirect UV detection: decoding of sun light from its light spectrum
Each light source has its characteristic light emission spectrum e.g. sun contains high red and blue light content compared to artificial light sources, such as tungsten light bulb have just high red and a very small fraction of blue light. Thus, we design a biological circuit which can distinguish between four different light conditions: i) high red-high blue; ii) high red-low blue; iii) low red-high blue and iv) low red-low blue (fig.). Such circuit consists of three hybrid promoters each acting as a NOR gate for two different transcriptional repressors i.e. transcription from such promoter is activated just when both repressors are absent.
We developed a novel photoinduction model to select for a suitable light receptors for the system inputs: LovTAP (blue light receptor) and Cph8 (Red light receptor). Later analysis of the circuit network revealed several kinetic parameters constrains upon which hybrid promoters were chosen and synthesized. Further testing of the promoters showed that as predicted, they act as a NOR gate, giving high potential of building a first full working light regulated biological decoder circuit in cells.
References
- Brown, B. a, Headland, L. R., & Jenkins, G. I. (2009). UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochemistry and photobiology, 85(5), 1147–55.
- Christie, J. M., Salomon, M., Nozue, K., Wada, M., & Briggs, W. R. (1999): LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proceedings of the National Academy of Sciences of the United States of America, 96(15), 8779–83.
- Christie, J. M., Arvai, A. S., Baxter, K. J., Heilmann, M., Pratt, A. J., O’Hara, A., Kelly, S. M., et al. (2012). Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science (New York, N.Y.), 335(6075), 1492–6.
- Cloix, C., & Jenkins, G. I. (2008). Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Molecular plant, 1(1), 118–28.
- Cox, R. S., Surette, M. G., & Elowitz, M. B. (2007). Programming gene expression with combinatorial promoters. Molecular systems biology, 3(145), 145. doi:10.1038/msb4100187
- Drepper, T., Eggert, T., Circolone, F., Heck, A., Krauss, U., Guterl, J.-K., Wendorff, M., et al. (2007). Reporter proteins for in vivo fluorescence without oxygen. Nature biotechnology, 25(4), 443–5
- Drepper, T., Krauss, U., & Berstenhorst, S. M. zu. (2011). Lights on and action! Controlling microbial gene expression by light. Applied microbiology, 23–40.
- EuropeanCommission (2006). SCIENTIFIC COMMITTEE ON CONSUMER PRODUCTS SCCP Opinion on Biological effects of ultraviolet radiation relevant to health with particular reference to sunbeds for cosmetic purposes.
- Elvidge, C. D., Keith, D. M., Tuttle, B. T., & Baugh, K. E. (2010). Spectral identification of lighting type and character. Sensors (Basel, Switzerland), 10(4), 3961–88.
- GarciaOjalvo, J., Elowitz, M. B., & Strogatz, S. H. (2004). Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proceedings of the National Academy of Sciences of the United States of America, 101(30), 10955–60.
- Gao Q, Garcia-Pichel F. (2011). Microbial ultraviolet sunscreens. Nat Rev Microbiol. 9(11):791-802.
- Goosen N, Moolenaar GF. (2008) Repair of UV damage in bacteria. DNA Repair (Amst).7(3):353-79.
- Heijde, M., & Ulm, R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends in plant science, 17(4), 230–7.
- Hirose, Y., Narikawa, R., Katayama, M., & Ikeuchi, M. (2010). Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proceedings of the National Academy of Sciences of the United States of America, 107(19), 8854–9.
- Hirose, Y., Shimada, T., Narikawa, R., Katayama, M., & Ikeuchi, M. (2008). Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proceedings of the National Academy of Sciences of the United States of America, 105(28), 9528–33.
- Kast, Asif-Ullah & Hilvert (1996) Tetrahedron Lett. 37, 2691 - 2694., Kast, Asif-Ullah, Jiang & Hilvert (1996) Proc. Natl. Acad. Sci. USA 93, 5043 - 5048
- Kiefer, J., Ebel, N., Schlücker, E., & Leipertz, A. (2010). Characterization of Escherichia coli suspensions using UV/Vis/NIR absorption spectroscopy. Analytical Methods, 9660. doi:10.1039/b9ay00185a
- Kinkhabwala, A., & Guet, C. C. (2008). Uncovering cis regulatory codes using synthetic promoter shuffling. PloS one, 3(4), e2030.
- Krebs in Deutschland 2005/2006. Häufigkeiten und Trends. 7. Auflage, 2010, Robert Koch-Institut (Hrsg) und die Gesellschaft der epidemiologischen Krebsregister in Deutschland e. V. (Hrsg). Berlin.
- Lamparter, T., Michael, N., Mittmann, F., & Esteban, B. (2002). Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proceedings of the National Academy of Sciences of the United States of America, 99(18), 11628–33.
- Levskaya, A. et al (2005). Engineering Escherichia coli to see light. Nature, 438(7067), 442.
- Mancinelli, A. (1986). Comparison of spectral properties of phytochromes from different preparations. Plant physiology, 82(4), 956–61.
- Nakasone, Y., Ono, T., Ishii, A., Masuda, S., & Terazima, M. (2007). Transient dimerization and conformational change of a BLUF protein: YcgF. Journal of the American Chemical Society, 129(22), 7028–35.
- Orth, P., & Schnappinger, D. (2000). Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nature structural biology, 215–219.
- Parkin, D.M., et al., Global cancer statistics, 2002. CA: a cancer journal for clinicians, 2005. 55(2): p. 74-108.
- Rajagopal, S., Key, J. M., Purcell, E. B., Boerema, D. J., & Moffat, K. (2004). Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochemistry and photobiology, 80(3), 542–7.
- Rizzini, L., Favory, J.-J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., Baumeister, R., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science (New York, N.Y.), 332(6025), 103–6.
- Roux, B., & Walsh, C. T. (1992). p-aminobenzoate synthesis in Escherichia coli: kinetic and mechanistic characterization of the amidotransferase PabA. Biochemistry, 31(30), 6904–10.
- Strickland, D. (2008). Light-activated DNA binding in a designed allosteric protein. Proceedings of the National Academy of Sciences of the United States of America, 105(31), 10709–10714.
- Sinha RP, Häder DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci. (2002). 1(4):225-36
- Sambandan DR, Ratner D. (2011). Sunscreens: an overview and update. J Am Acad Dermatol. 2011 Apr;64(4):748-58.
- Tabor, J. J., Levskaya, A., & Voigt, C. A. (2011). Multichromatic Control of Gene Expression in Escherichia coli. Journal of Molecular Biology, 405(2), 315–324.
- Thibodeaux, G., & Cowmeadow, R. (2009). A tetracycline repressor-based mammalian two-hybrid system to detect protein–protein interactions in vivo. Analytical biochemistry, 386(1), 129–131.
- Tschowri, N., & Busse, S. (2009). The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes & development, 522–534.
- Tschowri, N., Lindenberg, S., & Hengge, R. (2012). Molecular function and potential evolution of the biofilm-modulating blue light-signalling pathway of Escherichia coli. Molecular microbiology.
- Tyagi, A. (2009). Photodynamics of a flavin based blue-light regulated phosphodiesterase protein and its photoreceptor BLUF domain.
- Vainio, H. & Bianchini, F. (2001). IARC Handbooks of Cancer Prevention: Volume 5: Sunscreens. Oxford University Press, USA
- Quinlivan, Eoin P & Roje, Sanja & Basset, Gilles & Shachar-Hill, Yair & Gregory, Jesse F & Hanson, Andrew D. (2003). The folate precursor p-aminobenzoate is reversibly converted to its glucose ester in the plant cytosol. The Journal of biological chemistry, 278.
- van Thor, J. J., Borucki, B., Crielaard, W., Otto, H., Lamparter, T., Hughes, J., Hellingwerf, K. J., et al. (2001). Light-induced proton release and proton uptake reactions in the cyanobacterial phytochrome Cph1. Biochemistry, 40(38), 11460–71.
- Wegkamp A, van Oorschot W, de Vos WM, Smid EJ. (2007 )Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl Environ Microbiol. Apr;73(8):2673-81.
 "
"