Team:Peking/DataPage
From 2012.igem.org
How Our System Works
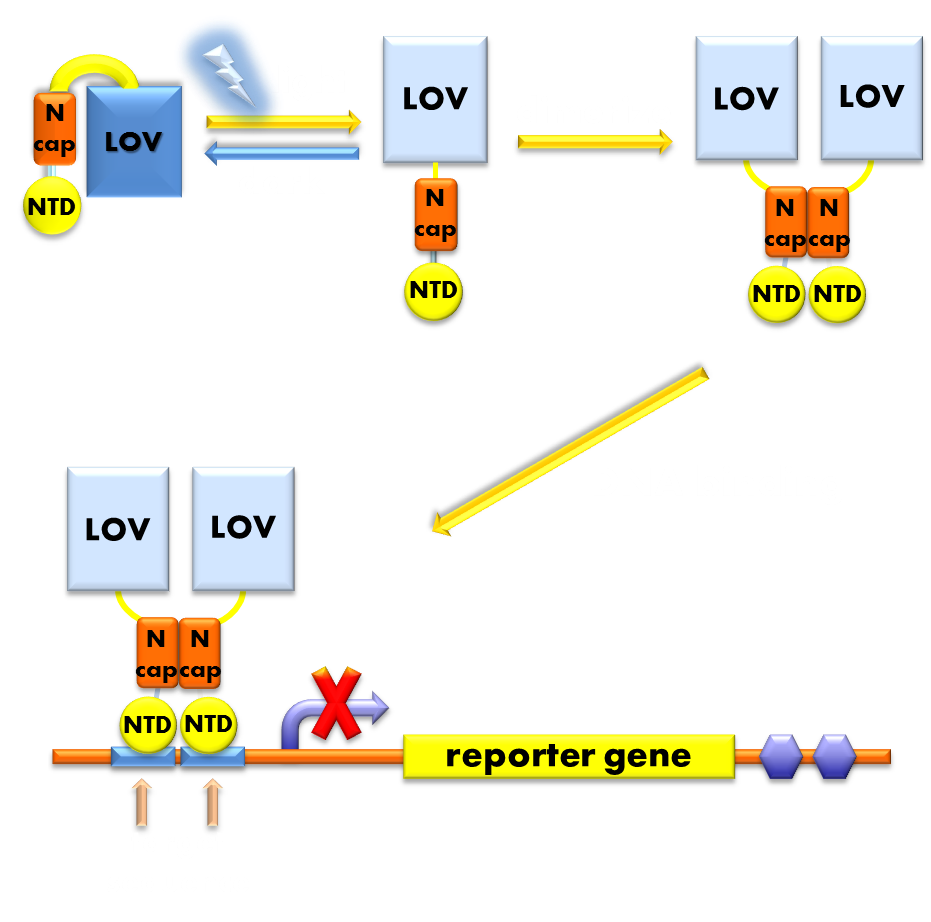
Figure 1. Illustration of how the Luminesensor functions. When exposed to blue light, the N-terminal cap of VVD domain will undock and cause VVD domain to dimerize. The dimerization of VVD protein will help the dimerization of DNA binding domain derived from the LexA protein. When helped to dimerize, DNA binding domain will recognize and bind to corresponding promoter, thus to inhibit transcription initiation.
Data for Favorite Parts
An Optimized Luminesensor(Main Page-BBa_K819005) A fusion protein consisting of DNA binding domain derived from E.coli endogenous SOS system repressor LexA, carrying mutations at positions 40-42, and a light-sensing domain from fungus N. crass photosensor protein VVD, carrying mutations N56K, C71V and I135M. This ultra-sensitive Luminesensor functions as a transcription repressor in response to blue light. Since we have introduced mutations into the LexA DNA binding domain, the Luminesensor proved orthogonal to host genetic context. Furthermore, we introduced mutations in the VVD photosensitive domain significantly improved the dynamic range of our optogenetic module.
Testing device for the Luminesensor (Main Page-BBa_K819006) and measurement device for the Luminesensor (Main Page-BBa_K819007) a fast degrading GFP ligated to the downstream of SulA408 promoter and RecA408 promoter, respectively. These two promoters are regulated by the Luminesensor and works independently of the host genetic context.
Data for Optimized Parts

Figure 2. BL21 cells harboring T7-lux operon(BBa_K819008) are emitting blue luminescence under IPTG induction at 5x10-5M
Lux Operon under T7 promoter (Main Page-BBa_K819008)
We successfully improved the easy-of-use of “lux brick” (BBa_K325909) part constructed by Cambridge 2010 iGEM team by placing the coding sequence of luxbrick (luxCDABEG) and related RBS under T7 promoter (BBa_I712074).
T7 promoter separates sensing/circuitry functions from pathways/actuation. It is encoded in genetically distinct regions while linked with other circuits by providing the output for the circuits that drive the expression of phage polymerases. Luxbrick under T7 promoter is very modular, because it is transcribed by T7 polymerase, which can be placed under any other promoter, forming a interface between luxbrick and other systems.
Once transformed into BL21(de3)strain, high protein production can be achieved rapidly after IPTG induction.Final concentration of IPTG should be round 0.05mM.
To note that the incubating temperature should be no higher than 30oC, or the heavy Lux complex can easily aggregate.
Optimum incubating conditions provided by Peking iGEM 2012:
250 rpm, 22oC, good ventilation after induction(final concentration of IPTG: round 0.5 mM).
Data for Other Submitted Parts
Luminesensor repressible SulA408 promoter (Main Page-BBa_K819017)
Luminesensor repressible RecA408 promoter (Main Page-BBa_K819002)
These are the two promoters used in this year’s project to test the functionality of Luminesensor and measure the efficiency of it.
Weak CheZ generator (Main Page-BBa_K819009)
medium CheZ generator (Main Page-BBa_K819010)
Chemotaxis protein CheZ ligated to 2 constitutive promoters with varying strengths. These two parts express the CheZ protein in varied quantities, which corresponds to the different levels of induced mobility in the ΔCheZ E.coli strain.
Luminesensor repressible RecA408 promoter (Main Page-BBa_K819015)
Luminesensor repressible SulA408 promoter (Main Page-BBa_K819016)
Chemotaxis protein CheZ ligated to the Luminesensor repressible SulA408 promoter and RecA408 promoter, respectively. Presumably these two parts can would convert chemotaxis pathway of E.coli into a phototaxis pathway by controlling the expression level of CheZ in ΔCheZ strain in response to blue light.
Luminesensor repressible LacIM generator (Main Page-BBa_K819012)
Coding sequence of LacI mutant protein driven by the Luminesensor repressible SulA408 promoter (BBa_K819017).
Data for Pre-Existing Parts
luxbrick (Main Page-BBa_K325909)
luxbrick is a bacteria luciferase part constructed by Cambridge 2010 iGEM team. They successfully expressed bacterial luciferase in E.coli, which could produce blue light under L-arabinose induction, and they carefully characterized this part. But because there are data we need while they didn’t give, we made the following supplementary characterization.
The supplementary characterization and our application of luxbrick part were added to the experience page of this part (see Experience-BBa_K325909)
The spectrum of the light emitted from E.coli harboring luxbrick was measured and it showed a maximum intensity at 485nm.

Figure 3. TOP10 cells harboring the luxbrick were cultured in LB medium and induced with L-arabinose at 10-3M. 10 hours after induction, the glowing cells were measured for spectrum using SHIMADZU RF5301PC Spectrofluorophotometer.
The time course of luxbrick expression and blue light emission under L-arabinose induction was given in a more visualized way, which is a short movie. As shown in the movie, the cells transformed with luxbrick begin to glow 9 hours after induction, and the entire visible glowing process lasts about 10 hours.
if you can not view this video,please click HERE.
Movie1. the time course of our light emitting cell
 "
"