Team:Technion/Project/Phage
From 2012.igem.org

Contents |
Objective
The main objective of this project is to create phage lambda that goes through its lytic cycle only under specific conditions that are met in the bacterial host. The idea is to replace one phage protein location in the genome, under a new regulatory promoter. This will allow the phage lytic cycle only in inducible conditions, controlled by the engineered plasmids that express RNA-polymerases.
This project included planning the genetic manipulation of the phage genome. This includes:
- Phage deviation into fragments that will ease the genetic manipulation, and re-factoring of the phages genome, after cutting it into fragments.
- Planning of the Q gene deletion and re-insertion under the desirable regulation, the RNA-polymerase promoter.
- The design of the antibiotic resistance gene insertion into the phage genome, in order to create additional selection to bacteria that contain the phage lysogenic genome.
The chosen phage lambda strain
The phage that was chosen as our working tool was phage lambda heat – inducible cI857s7. The phage genome contains four mutations:
- Addition of HindIII restriction site at 37,589 (C -> T) [ind1].
- Mutated S gene at 45,352 (G -> A), which leads to accumulation of infectious bacteriophage in the E. coli cells, the phage concentration increase when released from the cell [Sam7].
- Temperature sensitive mutation that converts the CI gene into a thermo sensitive protein. This allows inducing the lysogenic phage cycle in 37˚C, and lysis induction in 42˚C, this mutation is created at 37,742 (C -> T) [cI857].
- Additional point mutation at 43,082 (G -> A).
The phage genome sequence was taken from [http://www.ncbi.nlm.nih.gov/nuccore/NC_001416.1 NCBI]
The physical DNA was obtained from [http://www.neb.com/nebecomm/products/productn3011.asp NEB]
We chose to work with this phage mainly because of the temperature sensitivity, and the ability to induce lysis in controlled conditions. Moreover, the phage concentration will be higher when the bacterial cell undergoes lysis, due to the Sam7 mutation.
The division into fragments
When thinking on how to manipulate the phage genome and insert one or more genes under desire regulation (like RNA-pol promoters in our study), the first problem is how to manipulate a 48kb genome.
Our solution for this problem is to divide the phage genome into eight fragments.
Figure 1 shows the whole phage genome as divided into fragments.

The original figure was taken from: S.V. Rajagopala1, S. Casjens. and P. Uetz, 2011, "The protein interaction map of bacteriophage lambda", BMC Microbiology, 1, pp. 213-228.
Each fragment was inserted in the pSB1C3 plasmid, which was improved for our use by adding a MCS, with unique restriction sites that do not appear in the phage native genome ([http://partsregistry.org/Part:BBa_K784023 BBa_K784023]).
The main challenge with the phages genome division is to avoid putting regulatory elements with their regulated sequences. By doing so, we can prevent the expression of harmful and lytic associated proteins to the host bacteria. For example, all the lytic associated proteins are located together in fragment 8 (F8), without any promoter (PR) or activating protein (Q).
Moreover, due to the genome organization we can create function correlated sequences, each containing several proteins with a specific purpose. For example, the phage head proteins are located together in F1, and all the regulatory sequences are located in F6.
Results
pSB1C3+MCS
The MCS was constructed by DNA hybridization of two single stranded DNA molecules. The hybridized product was cloned into pSB1C3 using the XbaI and PstI1 sites. The MCS contains the following unique restriction sites: EcoRI, XbaI, BglII, HindIII, PacI, BamHI, SpeI and PstI.
To test the MCS, we digested 500ng of plasmid with each of the restriction enzymes. The restriction products were ran on a gel along with an uncut plasmid. The results are presented in Figure 2.
It can be noticed that all the enzymes cut efficiently except for XbaI. After looking again at the sequence of the MCS it was found that the combination of the overlapping XbaI and BglII sites created a GATC methylation site. XbaI digestion is affected by this methylation. Therefore, to achieve highest restriction efficiency with XbaI in this construct the plasmid should be propagated in dam- E. coli strains. A future improvement to this MCS would be restoring the original BioBrick prefix by inserting a G downstream to the XbaI site. This will destroy the BglII site but it will also destroy the GATC sequence, therefore, restoring the XbaI restriction site to a non methylated state.
Phage fragments
Each fragment was amplified using PCR reaction, with the phage lambda's genome as a template (The physical DNA was obtained from [http://www.neb.com/nebecomm/products/productn3011.asp NEB].
This procedure needed a fine tuning, because the amplification template was the phage's 48kb genome (which means more than one band for most fragments…). The primers for this reaction contained PacI and SpeI recognition sequences (see the primers sequences file). After gel purification, the products were restricted using PacI and SpeI, and ligated to the pSB1C3 improved plasmid [http://partsregistry.org/Part:BBa_K784023 BBa_K784023]. The ligation products were transformed into competent Top10-Rb (or Top10-Ca) bacteria, and were tested for the insert using colony PCR. Most of the fragments were longer than the plasmid backbone, the cloning wasn't as efficient as in shorter inserts.
We successfully cloned four out of the eight fragments.
The assembly strategy
Our next goal is to reassemble the eight fragments into the whole 48kb genome. We planned to use Gibson Assembly, with a few adjustments to the assembly of long fragments, since the standard Gibson Assembly solution we use is designed for shorter fragments.
Calibration of fragments overlap length and reassembly conditions
The goal of this experiment was to find the reaction conditions that allow the highest assembling efficiency when assembling long fragments.
Our first step was to assemble two fragments, while using different lengths of fragments' overlap and different concentrations of T5 exonuclease. We used 100bp, 250bp and 400bp overlap between F4 (8080bp) and F5 (6150bp).
The fragments with the different overlap length were amplified using PCR reaction, with the phage lambda genome as a template. The sense primers were identical to all the fragments, and the anti-sense primers were moved along the phage genome, in order to create the different overlaps (see the primers sequences file). This procedure was created using the NEB Gibson assembly buffer, with an addition of different concentrations of T5 exonuclease (1:100).
Results
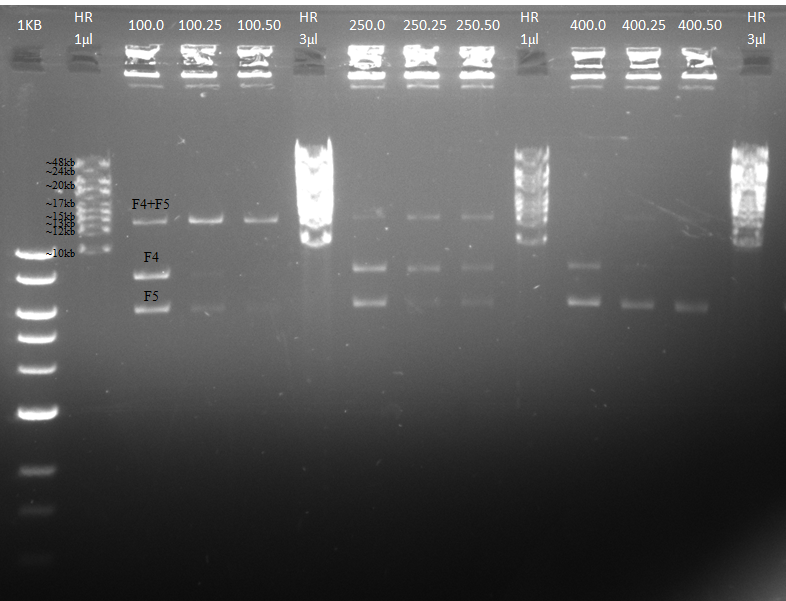
The T5 exonuclease increases the assembly efficiency for all fragments, with the most efficient reaction achieved in the 100bp fragment overlap with 0.5μl addition of T5 exonuclease.
All the reactions were performed with DNA weight of ~150ng for each fragment in a 5 μl volume. We added different T5 exonuclease concentration to each sample (0.25×104U and 50×104U per reaction) and the volume was completed to 10μl using NEB Gibson solution (5, 4.75 and 4.5 μl, in accordance to the T5-exo addition).
The results are described in Figure 3. We succeeded in assembling F4 (8080bp) and F5 (6150bp) into one fragment (14230bp), in all the overlaps lengths.
- When comparing lanes 1.0 (100bp overlap; no T5 addition), 2.0 (250bp overlap; no T5 addition) and 3.0 (400bp overlap; no T5 addition), it is visible that the 100bp fragment overlap reaction was the most efficient, and the 400bp was the least efficient.
- When comparing the three lanes at each fragments overlap length, it is clear that the addition of the T5 exonuclease increases the assembly efficiency for all fragments. In the 100bp fragments overlap the lower "source fragments" were hardly visible (lane 1.2), especially when compared to the control which didn't contain T5 exonuclease (lane 1.0).
- It is possible that an increase in the T5 exonuclease concentration, will allow the assembly for the 400bp fragments overlap.
Re-factoring the eight phage fragments together again
After we concluded that the 100bp fragments overlap yields the best fragments' assembly results, especially with the addition of T5 exonuclease, the next step is to assemble more than two fragments using these conditions. We chose to work with the following triple of fragments: F4 (8080bp), F5 (6150bp) and F6 (5700bp) and F5, F6 and F7 (7300bp), and the following four fragments: F4, F5, F6 and F7.

It can be seen that we succeeded in assembling two and three fragments. However, these reactions are not efficient as can be seen by the amount of DNA that was not assembled.
Results
All the reactions were performed in DNA weight of ~150ng for each fragment in a 5 μl volume. We added 50×104U T5 exonuclease per reaction and the volume was completed to 10μl using NEB Gibson solution. We created a reference sample using the "source fragment" as a ladder (marked in L). The expected length of the fragments described in Figure 4
From Figure 5, it can be seen that we succeeded in assembling two different fragments with low efficiency. The assembly of three and four fragments didn't go as well, since the bands that can be seen aren't very clear.
- We succeeded in assembling: F4+F5 (14230bp), F5+F6 (11850bp), as can be seen in the fourth lane from the left, F5+F6, and F4+F5, F5+F6, and F6+F7 (13000bp). However, these reactions' efficiencies were very low.
- Because the bands' intensity at the three and four fragments' assembly was low, we can conclude that the three fragments' reassembly succeeded, but with very low efficiency.
- By examining the "source bands" at all the reactions, it is visible that the reaction wasn't very efficient. It is possible that the DNA high concentration led to its collapse due to electrostatic forces. This could decrease the DNA availability to the enzymes, and therefore the assembly wasn't possible.
Our next experiment was a repetition of these reactions, using only three fragments, while testing the reaction time, the DNA concentration effect on the reaction efficiency, and the Gibson solution and T5 exonuclease concentrations. We found the reaction time and DNA concentrations didn’t affect the reaction efficiency.
In order to allow the entire eight phage lambda fragments assembly, we need to further examine the proper reaction conditions. Our vision for the next steps is: "1, 2, ∞", and we hope that if we succeeded in the two and three fragments assembly, we will successfully reassemble all of the eight phage fragments together again.
Strategy for the antibiotic resistance addition
We planned to insert Kanamycin antibiotic resistance gene to the phage genome. The purpose of the resistance gene insertion is to create additional selection to bacteria containing the lysogenic phage. The phage can be used as an easy tool for inserting large cassettes into bacteria, after deleting its lytic fragment and keeping the site specific recombination fragment.
We planned to take the antibiotics resistance gene from pSB1K3 backbone with a constitutive promoter and to insert it in the middle of fragment 6, between the PL promoter and the rexA gene (shown in Figure 1). This will allow the insertion of the resistance gene, while the phage reading frames won't be damaged.
The resistance gene will be inserted to the phage using Gibson assembly of three fragments: Kan resistance sequence that will contain two overlaps to F6 "right arm" and to F6 "left arm". The cassette structure is described in Figure 6.
The Q gene
The Q gene is one of the two main regulatory proteins (Q and N), which regulates the expression of the lytic proteins (S and R). We chose to work with the Q gene because it is the main regulator in the phage lytic cycle, in the absence of it – the phage won't be able to cause lysis. Therefore, we expect not to see any plaques on the growth plate in the phage with the deleted Q gene. Our next goal is to delete the Q gene from fragment 7 ([http://partsregistry.org/Part:BBa_K784021 BBa_K784021]) using deletion PCR, and re-insert it into a different position in the phage genome. The plan was to insert it back in fragment 4, between the tail fiber proteins' coding sequence and the b2 region (shown in Figure 1). This is the region between the early left operon (sequence transcription starts with the PL promoter and regulated by N gene) and the late operon (sequence transcription starts from the PR and regulated by Q gene). The two operons are in opposite notations. The structure of fragment 7 after the Q gene deletion, and fragment 4 after the Q gene insertion, is describes in Figure 7.
As mentioned the chosen Q gene will be reinserted with a specific promoter that will allow the genes' transcription and translation only in a bacteria that will express the promoter's activator (RNA-pol in our work plan).
References
- Gibson D. G., et al. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6(5): 343-345.
- Gottesman M. E., Weisberg R. A. 2004. Little Lambda, Who Made Thee? Microbiology and Molecular Biology Reviews 68(4): 796-813.
- Rajagopala S., Casjens S., Uetz P. 2011. The protein interaction map of bacteriophage lambda. BMC Microbiology 11(1).
 "
"



