Team:Calgary/Notebook/Electrochem
From 2012.igem.org
| Line 21: | Line 21: | ||
<p>Results from the model have been obtained, hinting at a detection time of approximately 250 seconds, or just over 4 minutes. This is based on the detection of PAP at 0.06mM, which is equivalent to the 1500 μL of PAP sweep shown in Figure 2. The sequencing results from the <i>lacZ</i> and <i>uidA</i> genes have arrived and the <i>lacZ</i> construct is fine while the <i>uidA</i> construct has some mutations.</p> | <p>Results from the model have been obtained, hinting at a detection time of approximately 250 seconds, or just over 4 minutes. This is based on the detection of PAP at 0.06mM, which is equivalent to the 1500 μL of PAP sweep shown in Figure 2. The sequencing results from the <i>lacZ</i> and <i>uidA</i> genes have arrived and the <i>lacZ</i> construct is fine while the <i>uidA</i> construct has some mutations.</p> | ||
<h2>Week 10 (July 3-6)</h2> | <h2>Week 10 (July 3-6)</h2> | ||
| - | <p>A standard curve was attempted for PAP, however the graphs recorded resembled a solution of intense resistivity. A possible explanation for this is the clip connecting the potentiostat to the electrode having an incomplete connection. An electrode with a stronger connection | + | <p>A standard curve was attempted for PAP, however the graphs recorded resembled a solution of intense resistivity. A possible explanation for this is the clip connecting the potentiostat to the electrode having an incomplete connection. An electrode with a stronger connection was constructed and tested to remedy this, however it presented no detection of the PAP at any concentration. Switching to a platinum working electrode showed a strong response and testing will continue with this next week.</p> |
</html> | </html> | ||
Revision as of 16:23, 10 July 2012
Week 1 (May 1-4)
This week involved planning various experiments and gathering chemicals to be tested. Unfortunately no lab work was conducted this week.
Week 2 (May 7-11)
Chlorophenol red (CPR) was oxidized in a sodium phosphate solution with zinc phthalocyanine modified carbon electrodes or just straight carbon electrodes. It was found that the modified electrodes had greater responses than their unmodified counterparts. All future work with CPR will be done with these electrodes. An IPTG induced lacZ was transformed for the registry and the uidA gene was amplified and biobricked under the control of the IPTG promoter. These genes will be used to cleave specific sugars from electrochemical analytes.
Week 3 (May 14-18)
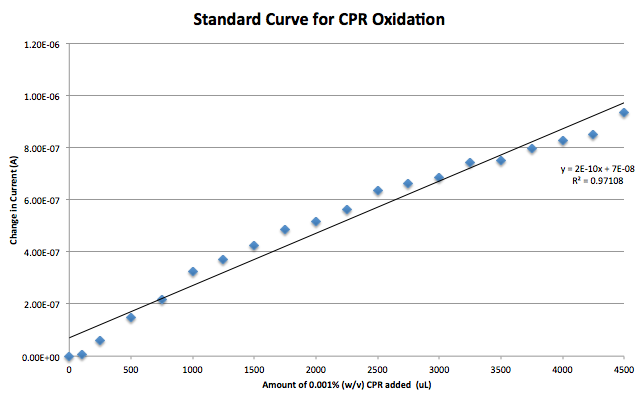
A standard curve was generated using CPR as the analyte in a sodium phosphate solution. This was made with the modified electrodes. After this it was found that the reference electrode being used, a silver/ silver chloride electrode, had stopped functioning. Switching to a reduction of hydrogen electrode (RHE) reference fixed this problem. A gold working electrode was also briefly tested to reduce the background noise caused by the capacitance of carbon. Further testing will be needed to chose a final electrode.
Week 4 (May 21-25)
This week involved testing the oxidation of CPR and para-aminophenol (PAP) in the same solution to determine if their oxidation potentials were unique. By showing that they are separate peaks it demonstrates that they can be used as two of the electrochemical components of a multiplexed biosensor. After a morning of failed tests on Thursday I finally managed to show that the peaks are unique (0.7V for PAP and 1.3V for CPR vs RHE), giving us the first step towards a final sensor.

Week 5 (May 28- June 1)
This week the data collected was inconsistent across multiple runs even though the electrodes used were from the same batch. It seems that each of these electrodes is unique, presenting a challenge for analyzing results when multiple electrode setups are used.
Week 6 (June 4-8)
This week marked the start of the construction of a mathematical model of the electrochemical system. The goal of this model is to identify the shortest time in which an electrochemical signal could be reached and what conditions would decrease this detection time. To do this the MATLAB toolbox SimBiology will be used as well as differential solvers and published rate constants. The model will include translation, transcription, RNA and protein degradation, and the actual conversion from the sugar conjugate into the electrochemical analyte.
Week 7 (June 11-15)
The list of final analytes was compiled this week, giving a goal of chemicals to test with and without their sugar conjugates. The genes necessary to break these compounds apart were also noted and primers were designed so that they could be amplified. More work was placed on the model as well, getting it to the point where it can go from a constant amount of plasmid DNA into the RNA transcript and then the protein product.
Week 8 (June 18-22)
The model has been updated to include the transport of PAPG, the sugar conjugated form of PAP, into the cell through the LacY transport protein. The diffusion of PAP across the membrane out into the solution was also included into the model. A diagramatic representation of this is shown below. The last of the chemicals needed for the electrochemical testing was ordered this week too, meaning that work can begin at an increased pace on the wetlab front when the shipment arrives.
Week 9 (June 25-29)
Results from the model have been obtained, hinting at a detection time of approximately 250 seconds, or just over 4 minutes. This is based on the detection of PAP at 0.06mM, which is equivalent to the 1500 μL of PAP sweep shown in Figure 2. The sequencing results from the lacZ and uidA genes have arrived and the lacZ construct is fine while the uidA construct has some mutations.
Week 10 (July 3-6)
A standard curve was attempted for PAP, however the graphs recorded resembled a solution of intense resistivity. A possible explanation for this is the clip connecting the potentiostat to the electrode having an incomplete connection. An electrode with a stronger connection was constructed and tested to remedy this, however it presented no detection of the PAP at any concentration. Switching to a platinum working electrode showed a strong response and testing will continue with this next week.
 "
"