Team:Calgary/Project/HumanPractices/Killswitch/Regulation
From 2012.igem.org
| Line 8: | Line 8: | ||
<h1>Magnesium repressible system</h1> | <h1>Magnesium repressible system</h1> | ||
| - | This system is repressed in the presence of magnesium. This system has two control components – a promoter and a riboswitch. Normally the magnesium promoter (mgtA promoter) and the magnesium riboswitch (mgtArb) are activated if there is a deficiency of magnesium in the cell. The lack of magnesium activates other genes in <i>E. coli </i>to allow influx of magnesium into the cell. There are two proteins in the cascade that activate the system namely PhoP and PhoQ. PhoQ is the trans-membrane protein which gets activated in the absence of magnesium and phosphorylates PhoP. PhoP in turn binds to the mgtA promoter and transcribes genes downstream.</p> | + | <p>This system is repressed in the presence of magnesium. This system has two control components – a promoter and a riboswitch. Normally the magnesium promoter (mgtA promoter) and the magnesium riboswitch (mgtArb) are activated if there is a deficiency of magnesium in the cell. The lack of magnesium activates other genes in <i>E. coli </i>to allow influx of magnesium into the cell. There are two proteins in the cascade that activate the system namely PhoP and PhoQ. PhoQ is the trans-membrane protein which gets activated in the absence of magnesium and phosphorylates PhoP. PhoP in turn binds to the mgtA promoter and transcribes genes downstream.</p> |
</html> | </html> | ||
| Line 39: | Line 39: | ||
</html> | </html> | ||
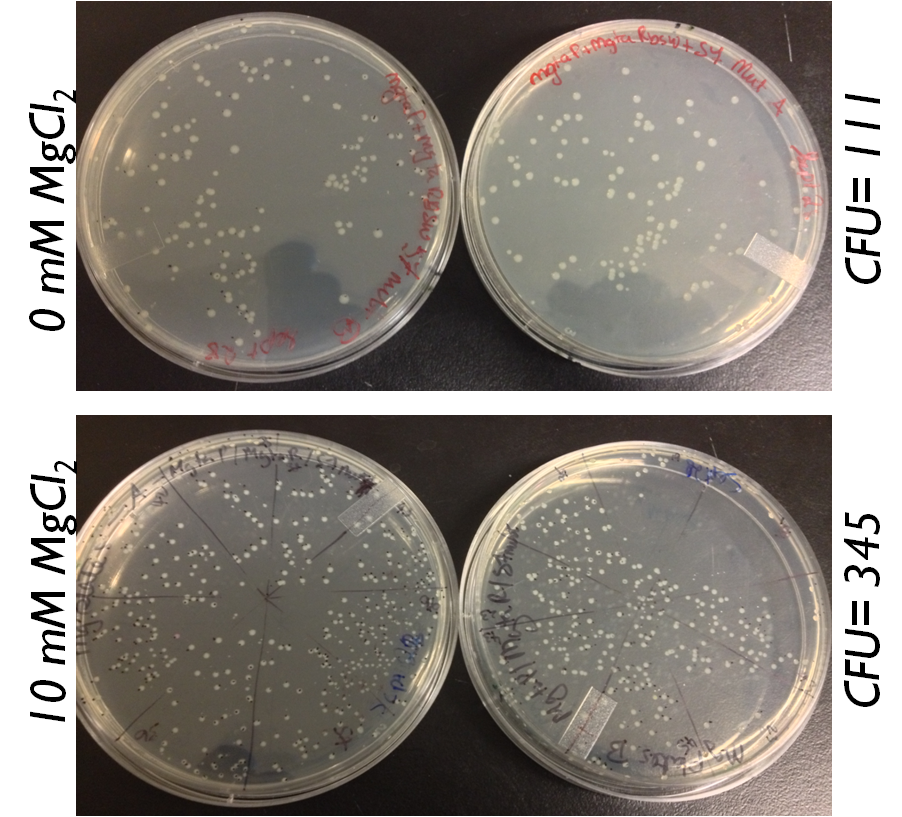
[[File:S7 mutagenesis for the magnesium system ucalgary.png|thumb|400px|center|Figure : Plating of S7 mutagenesis on plates with both magnesium and without magnesium. The plate with magnesium has more colonies compared to the plates without magensium.]]<html> | [[File:S7 mutagenesis for the magnesium system ucalgary.png|thumb|400px|center|Figure : Plating of S7 mutagenesis on plates with both magnesium and without magnesium. The plate with magnesium has more colonies compared to the plates without magensium.]]<html> | ||
| - | |||
| - | |||
<h1>Manganese regulation</h1> | <h1>Manganese regulation</h1> | ||
Revision as of 01:44, 2 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Regulation/Expression Platform
Magnesium repressible system
This system is repressed in the presence of magnesium. This system has two control components – a promoter and a riboswitch. Normally the magnesium promoter (mgtA promoter) and the magnesium riboswitch (mgtArb) are activated if there is a deficiency of magnesium in the cell. The lack of magnesium activates other genes in E. coli to allow influx of magnesium into the cell. There are two proteins in the cascade that activate the system namely PhoP and PhoQ. PhoQ is the trans-membrane protein which gets activated in the absence of magnesium and phosphorylates PhoP. PhoP in turn binds to the mgtA promoter and transcribes genes downstream.

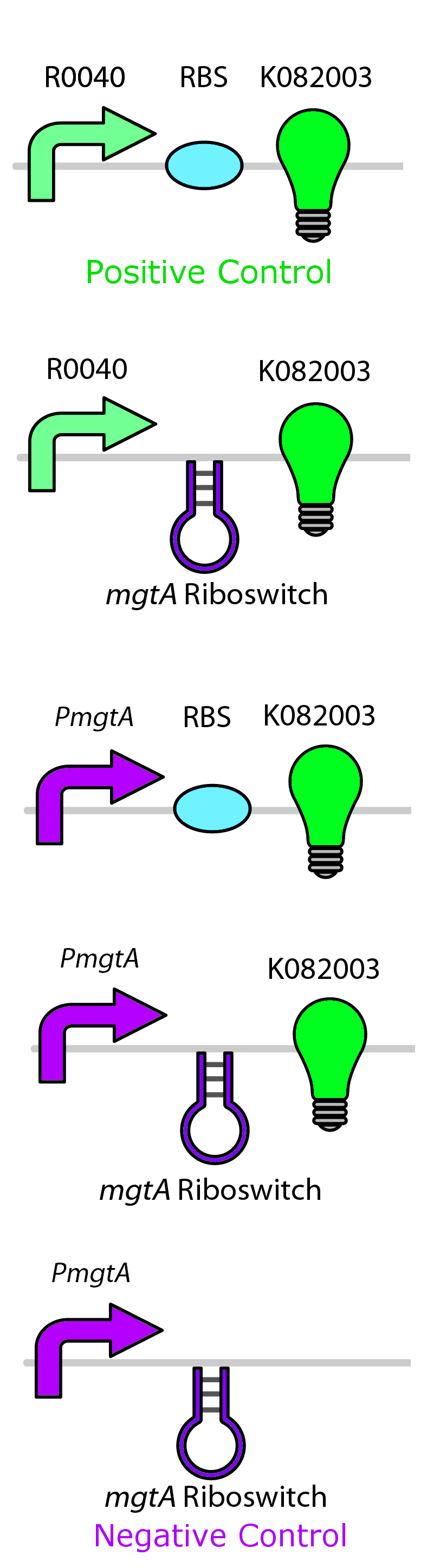
Test circuits for the magnesium system
To test the magnesium regulatory elements we built each of the elements with a reporter gene. We chose Bba_K082003 which is GFP with an LVA tag as our choice of reporter. We did not choose BBa_E0040, the stable GFP, because we wanted a real time indication of the system's control. Stable GFP has a half life of 8 hours and would still fluoresce when the system is shut off.
We build these circuits to test the control elements of the system, namely the mgtA promoter and the mgtA riboswitch.
Characterization of these circuits
We tested the aforementioned circuits in different concentrations of magnesium. For detailed protocol see INSERT LINK HERE. The values were normalized to the negative control which is the magnesium promoter and riboswitch alone.

There is a much larger drop in the GFP output when the mgtA promoter and riboswitch are working together compared to the mgtA riboswitch under the control of TetR promoter. This suggests that having both the promoter and the riboswitch together provides a tighter control over the genes expressed downstream. This also suggests that magnesium riboswitch alone is sufficient in inhibiting gene expression downstream of a constitutive promoter.
It is important to consider however that the control elements of the system namely PhoP and PhoQ were not present in the circuits tested. We believe that would give us much better control. Although the data suggests that there is enough production of PhoP and PhoQ proteins from the genome to control expression of the genes downstream of a high copy plasmid.
Although the magnesium system is a brilliant system which is highly regulated, it is not a suitable system for the purposes of our bioreactor. The tailings are composed of very high concentration of magnesium- upto 30mM(REFERENCE). As can be seen from figure 3, this would inhibit the system. Therefore, if our bacteria escapes into the tailings, the kill genes would not be activated and the bacteria would be able to survive.
In contrast, it is important to note that this system adds important regulatory elements to the registry such as an inducible promoter and a riboswitch which can be used by other teams to control both killswitches as well as other regulatory pathways which do not pertain using tailings.
We also wanted to test the magnesium system with our kill genes. The micrococcal nuclease that arrived from IDT had 1bp mutation which changed a isoleucine residue into a lysine. Therefore we had to mutate it. We strategized it such that we built our circuits with S7 first and then mutated the S7. We plated the mutated plasmid on plates with magnesium and without magnesium.
Manganese regulation
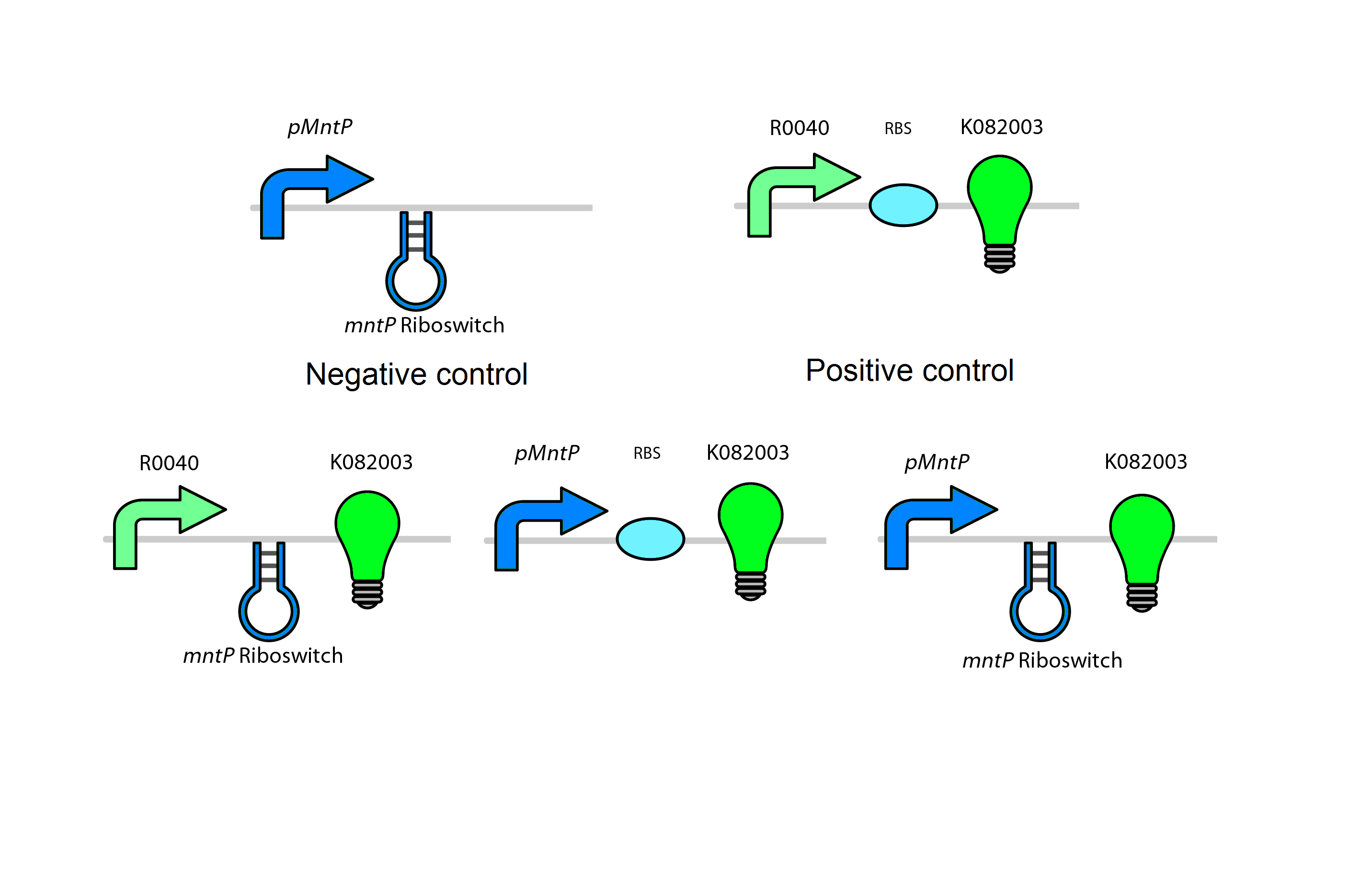
Manganese is found to be an essential nutrient in most bacteria and larger organisms. Due to the properties of manganese this metal can work as a catalyst for many chemical reactions in the body of organisms as well as playing a key role in the structure of macromolecules. For the use in our system, manganese will work as a response mechanism that will trigger the effects of this system. Similar to the designs created for the magnesium system, the manganese system will focus mainly on both the promoter and the riboswitch (mntP Promoter and mntP Riboswitch). Besides the promoter and riboswitch, this system also contains a transcriptional regulator (mntR). This metalloregulatory protein is found to observe the level of manganese and responds accordingly. The structure of this protein consists of a metal-binding domain that will respond in the presence of manganese and repress the manganese ion transporter (mntH). When this protein membrane is repress the bacteria cannot gained the needed manganese for survival. It is also found that the mntR positively regulates the mntP promoter which in turn up-regulates our manganese system. The mntP promoter in the presence of manganese initiates the transcription of mntP riboswitch which responds in a reciprocal manner compared to the magnesium system. Instead of ending transcription when the metal is accompanied, the mntP riboswitch will initiate when accompany by manganese and express the kill genes S7 and CViAII downstream of it.
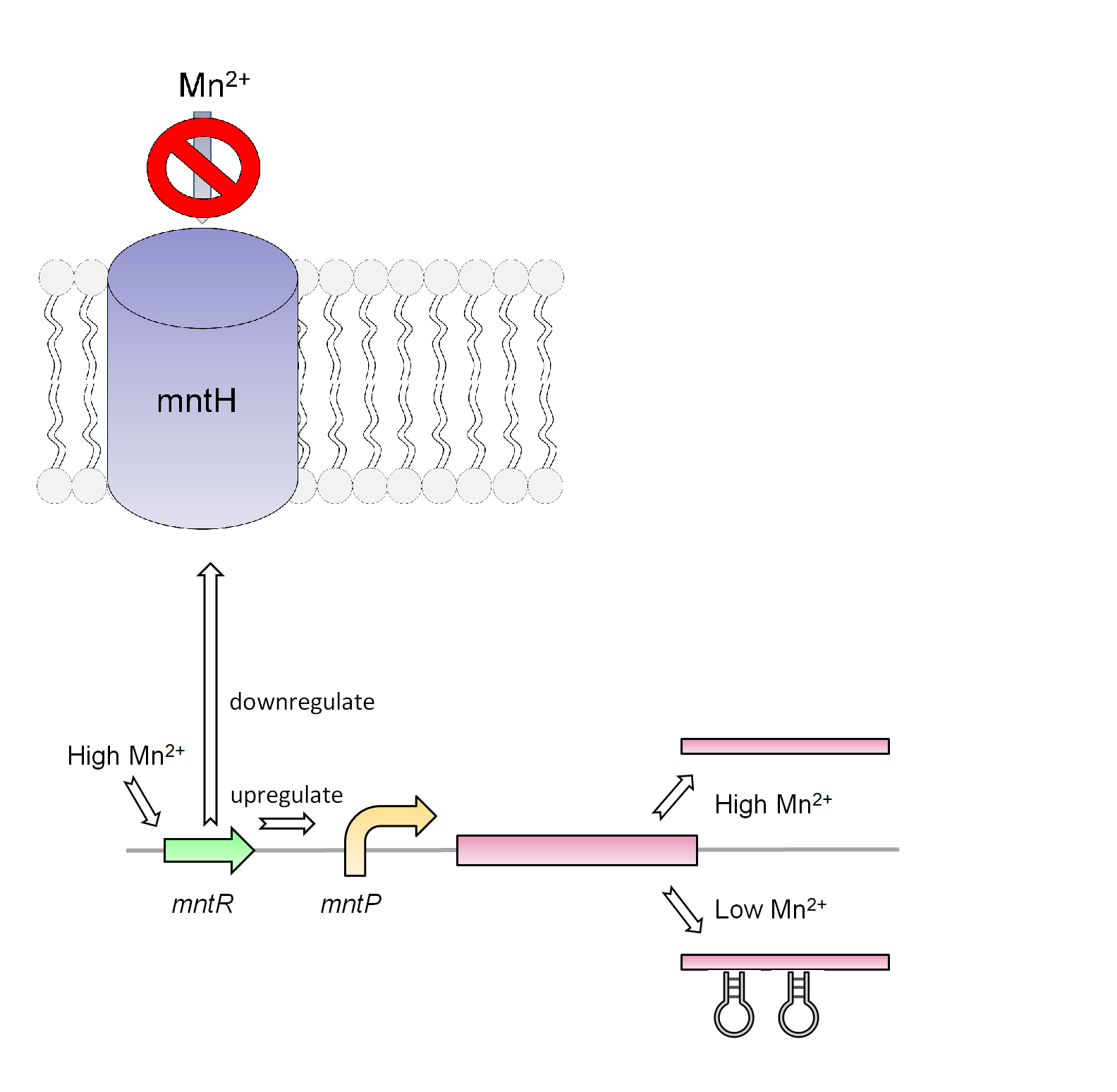
Both the magnesium and manganese systems are both workable killswitch constructs however after analyzing the composition of the tailing ponds the systems will not be viable without a way to regulate these two metals in the contaminated waters.
Test circuits for the manganese system
Similar to the magnesium system the manganese will use a GFP LVA tag. The following are the control circuits built in order to characterise the MntA promoter and the MntA riboswitch.
Glucose repressible system
Background
Given that oil sands tailings ponds contain significant amounts of magnesium and low levels manganese, and that the MgtA and MntP expression platforms are repressed by these conditions respectively, these systems are not appropriate for the bioreactor in a typical tailings pond site (FIND PAPER WHICH CITES IONs in TP). In order to ensure that the kill genes would be activated should bacteria escape from the bioreactor, we required a control element which would be expressed under conditions in typical tailings ponds. To this end, we selected a rhamnose inducible promoter from Eschericia coli as a potential method for regulating our kill gene combination.
The rhamnose inducible system is optimal in the bioreactor since the promoter is tightly repressed with the presence of glucose. We aim to supplement the bioreactor with low levels of glucose so that the kill genes downstream of the promoter would be repressed. In the event of escape into the tailings ponds, glucose levels would be insufficient for repression of the system, which would thus activate expression of the kill genes.
Although our team has also characterized a molydenum-repressed MOCO riboswitch as an additional control mechanism suitable to the bioreactor, we investigated the rhamnose promoter because of the lower cost and availability of the repression agent.
Native function of the rhamnose promoter
The rhamnose promoter (pRha) is responsible for regulating six genes related to rhamnose metabolism and contains a separate promoter on its leading and reverse strands (see Figure one). RhaR and RhaS are downstream on one side of the promoter and are the control proteins which regulate expression of the RhaB, RhaA, and RhaD genes on the opposite side of the promoter. Basal levels of RhaR transcription factor are activated by complexing with L-rhamnose so that expression of the rhaSR operon is up-regulated. In turn, the resulting RhaS activates the rhaBAD operon (Egan & Schleif, 1993). The RhaB, RhaA, and RhaD genes are directly involved in metabolism of rhamnose.
Although RhaS acts directly on pRha, Egan and Schleif (1993) proposed that extent of up-regulation of rhaBAD is dependent on two other factors: firstly, RhaS is more efficient when cAMP receptor protein (CRP-cAMP) is bound to the promoter; and secondly, RhaS causes higher expression of rhaBAD in the presence of rhamnose (see figure two).
Via the CRP-cAMP complex, glucose represses the rhaBAD operon through Eschericia coli's system of global catabolite represssion. In the presence glucose, membrane bound EIIA protein transfers phosphate to glucose. Under this condition, desphosphorylated EIIA is unable to activate adenyl cyclase resulting in lower levels of cAMP (CITE NATURE ARTICLE). In-turn, catabolite receptor protein (CRP) is unable to complex with cAMP, causing a down-regulation of the rhaBAD operon (see figure two, figure three (of CRP sites on promoter)).
Engineering pRha into a kill system
Our team has engineered the following rhamnose promoter kill system:
(Draw the final kill circuit)
In place of the rhaBAD operon, we have placed the CviAII endonuclease and S7 exoendonuclease for the active components of our kill system. We are capitalizing on glucose's global catabolite repression of these two genes as the controlled condition to repress cell destruction in the bioreactor. The rhamnose promoter as tightly controllable expression platform was inspired by two papers.
Giacalone et al. (2006) cloned the rhamnose promoter together with the rhaSR operon and proposed pRha as a viable system for expressing toxic proteins in E. coli. They tested the system in three different plasmids of varying copy number and replaced rhaBAD operon and characterized the system with GFP and TphoA protein. In their results, they found that 0.2% w/v D-glucose was significantly repressed GFP in the presence of L-rhamnose, and that it was completely repressed when only glucose was present. Likewise, they found that basal expression of TphoA was completely repressed when glucose was present in 0.2% w/v.
Jeske and Altenbuchner (2010) assembled a similar system with pRha, rhaSR operon, and GFP and found that florescence output was twenty-four times greater with rhamnose as opposed to rhamnose and glucose.
In designing this system for the bioreactor, we set out to replicate this tight glucose repression of the rhamnose promoter. As opposed to Giacalone et al. (2006) and Jeske and Altenbuchner (2010), we manipulated the rhaSR operon to better suit conditions in the bioreactor (see figure X).
The logic behind these changes is that rhamnose will not be present in the tailings ponds to activate the rhaSR operon cascade for induction of the promoter. Expression of these control genes are dependent on the rhamnose activation of RhaR. Given that rhamnose is not naturally present in tailings ponds, and that we are depending on the native composition of the waste water to activate the kill system, we had to modify the control system.
RhaS is the protein which directly activates the rhaBAD operon side of the pRha promoter. To bypass the natural induction by rhamnose, we put rhaS under control of a constitutive promoter (R0040) from the parts registry. In doing so, we intend to elevate expression of RhaS such that it will be able to activate pRha. When glucose levels are low, as is the case of the environment outside of the bioreactor, cAMP-CRP complexes bind to pRha so that the over-expressed RhaS may initiate the kill system.
Some may object to this system because Egan and Schleif (1993) suggested that RhaS was more efficient at activating pRha in the presence of rhamnose. This does not invalidate the methodology of our system though because Egan and Schleif (1993) only suggested that function of RhaS was optimized in the presence of rhamnose. They agreed that RhaS significantly upregulated pRha even when rhamnose was not available.
Assembly methods
For the construction of this system, our team had pRha commercially synthesized as it was described in Jeske and Altenbuchner (2010). SHOW PIC MAYBE
Additionally, we amplified rhaS and rhaR from Top Ten E. coli using Kapa Hi-Fi polymerase and PCR. Here follows the parts which we submitted for this system: (LIST THE PARTS)
Characterization
We followed similar protocols for characterizing pRha as Jeske and Altenbuchner (2010). In place of the rhaBAD operon, we inserted a gene for GFP with an LVA degradation tag. ASK WHAT WE DO REGARDING PROTOCOLS.
 "
"