Team:UTP-Software/SoftwareTool
From 2012.igem.org
(→Next S2MT Version) |
|||
| Line 15: | Line 15: | ||
<li> And last, if the program successfully created the required primer, it will print the sequence, giving the user the start position where that primer should be applied. </li> | <li> And last, if the program successfully created the required primer, it will print the sequence, giving the user the start position where that primer should be applied. </li> | ||
In some cases, it’s impossible to fix some sequences, because based on the protocol, the output primer may not comply with the steps that are listed in the “Protocol Followed to design the primers” section. Meaning that another standard should be selected. </ul></div> | In some cases, it’s impossible to fix some sequences, because based on the protocol, the output primer may not comply with the steps that are listed in the “Protocol Followed to design the primers” section. Meaning that another standard should be selected. </ul></div> | ||
| + | |||
| + | ==== Flow diagram for the S<sup>2</sup>MT ==== | ||
| + | |||
| + | [[File:Flow diagram.png|center]] | ||
| + | |||
==== Protocol Followed to design the primers:<sup>[http://web.physics.ucsb.edu/~deborah/pro/pro_pdf/Stratagene%20QuikChange.pdf]</sup> ==== | ==== Protocol Followed to design the primers:<sup>[http://web.physics.ucsb.edu/~deborah/pro/pro_pdf/Stratagene%20QuikChange.pdf]</sup> ==== | ||
| Line 25: | Line 30: | ||
<li> Primers need not be 5´ phosphorylated but must be purified either by fast polynucleotide liquid chromatography (FPLC) or by polyacrylamide gel electrophoresis (PAGE). Failure to purify the primers results in a significant decrease in mutation efficiency.</li> | <li> Primers need not be 5´ phosphorylated but must be purified either by fast polynucleotide liquid chromatography (FPLC) or by polyacrylamide gel electrophoresis (PAGE). Failure to purify the primers results in a significant decrease in mutation efficiency.</li> | ||
<li> It is important to keep primer concentration in excess. Stratagene suggests varying the amount of template while keeping the concentration of the primer constantly in excess.</li></ul></div> | <li> It is important to keep primer concentration in excess. Stratagene suggests varying the amount of template while keeping the concentration of the primer constantly in excess.</li></ul></div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
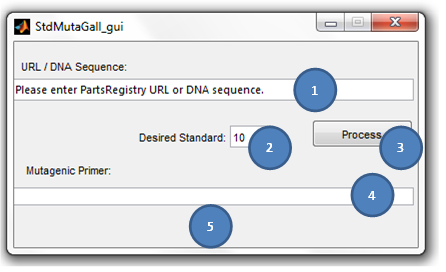
== '''S<sup>2</sup>MT Tutorial''' == | == '''S<sup>2</sup>MT Tutorial''' == | ||
<div align="justify"><ul> | <div align="justify"><ul> | ||
| - | [[File:Img1.png]] | + | [[File:Img1.png | center]] |
| Line 60: | Line 55: | ||
<div align="justify"><ul> | <div align="justify"><ul> | ||
| - | [[File:Fix.png]] | + | [[File:Fix.png |center]] |
(S<sup>2</sup>MT) succesfully fixing the incompatibility of the given DNA sequence with the standard 21. | (S<sup>2</sup>MT) succesfully fixing the incompatibility of the given DNA sequence with the standard 21. | ||
Revision as of 03:02, 27 September 2012
| Home | Team & Attributions | Project | S2MT | Tutorial | Biosinergia | Notebook | Human Practices | Safety | Sponsors |
|---|
|
|
Project DetailsWhat is the S2MT tool?
The SynBio Sequence Mutation Tool (S2MT) it's a MATLAB implementation software that helps teams to start their projects once they have selected some possible BioBricks or DNA sequences to work with. This tool was developed in MATLAB basically for the reason that, what new iGEM teams require is a flexible environment capable of not only running the algorithm that we develop, but also a place where they can continue working their ideas, examine more in detail their sequences and even continue the development of our tool. So we thing that our system will also knock down the barriers that exist between computation and biology and give students the tools required to do better simulations, and save a lot of valuable time! Behind the code there are three simple steps in which the program consists: Flow diagram for the S2MT
Protocol Followed to design the primers:[http://web.physics.ucsb.edu/~deborah/pro/pro_pdf/Stratagene%20QuikChange.pdf]
Tm = 81.5 + 0.41(%GC) − 675 / N - where N is the primer length in base pairs. S2MT Tutorial
Some points to take in consideration:
(S2MT) succesfully fixing the incompatibility of the given DNA sequence with the standard 21.
To download the script go to: https://github.com/igemsoftware/UTP_Software_2012 and get all three files. Files descriptions: Note that (S2MT) relies on the Bioinformatics Toolbox of MATLAB (both supplied by iGEM) to make its computations, so you need the toolbox to be able to run the script. Next S2MT VersionAfter finishing the first version, we had in mind several things to implement to the next:
One of them may be the Gibson Assembly[http://openwetware.org/wiki/Prather:Gibson_CBA] standard, which still requires compatibility with the actual methods, but include several advantages as: References[1]. Stratagene. QuikChange™ Site-Directed Mutagenesis Kit. Catalog #200518. Revision #108005h. Available in: http://web.physics.ucsb.edu/~deborah/pro/pro_pdf/Stratagene%20QuikChange.pdf. [2]. Prather:Gibson CBA. Available in: http://openwetware.org/wiki/Prather:Gibson_CBA |
 "
"