Team:Technion/Project/YES gates
From 2012.igem.org
(→References) |
(→YES gate #3) |
||
| Line 33: | Line 33: | ||
==YES gate #3== | ==YES gate #3== | ||
The third YES gate consists of the P<sub>Lux</sub> promoter and a [https://2012.igem.org/Team:Technion/Project/RS theophylline induced riboswitch (RS)]. In this YES gate, there is actually a two step regulation. The transcription from the P<sub>Lux</sub> promoter is induced by [http://partsregistry.org/3OC6HSL 3OC<sub>6</sub>HSL]. The translation is regulated by the RS and is induced by theophylline.<br> | The third YES gate consists of the P<sub>Lux</sub> promoter and a [https://2012.igem.org/Team:Technion/Project/RS theophylline induced riboswitch (RS)]. In this YES gate, there is actually a two step regulation. The transcription from the P<sub>Lux</sub> promoter is induced by [http://partsregistry.org/3OC6HSL 3OC<sub>6</sub>HSL]. The translation is regulated by the RS and is induced by theophylline.<br> | ||
| - | The RS was originally developed in fusion to DsRed Express {1}, which is a red FP from which mCherry was engineered {2}. Since the coding region has effect on the RS function, we thought mCherry would be a good candidate. Unfortunately, this turned out to be a bad choice, since the regulation by the riboswitch failed. The chronicles of the cloning and experimentation with the RS are described in the [https://2012.igem.org/Team:Technion/Project/RS RS] section. | + | The RS was originally developed in fusion to DsRed Express {1}, which is a red FP from which mCherry was engineered {2}. Since the coding region has effect on the RS function, we thought mCherry would be a good candidate. Unfortunately, this turned out to be a bad choice, since the regulation by the riboswitch failed. The chronicles of the cloning and experimentation with the RS are described in the [https://2012.igem.org/Team:Technion/Project/RS RS] section.<br> |
| + | Since the RS regulates the translation of a protein that is fused downstream to it, it had to be fused not only to the FP but also to the RNAPs. This step was completed successfully for several RNAPs as described in the [https://2012.igem.org/Team:Technion/Project/RS RS] section. | ||
==References== | ==References== | ||
1. <b>Lynch S. A., Gallivan J. P.</b> 2009. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Research <b>37</b>(1): 184-192.<br> | 1. <b>Lynch S. A., Gallivan J. P.</b> 2009. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Research <b>37</b>(1): 184-192.<br> | ||
2. <b>Shaner N. C., et al.</b> 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol <b>22</b>(12): 1567-1572. | 2. <b>Shaner N. C., et al.</b> 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol <b>22</b>(12): 1567-1572. | ||
Revision as of 16:59, 23 September 2012

Contents |
What's a YES gate?
A yes gate is one of the simplest molecular logic gate operations. The truth table for a YES gate is:
| Input | Output |
|---|---|
| 1 | 1 |
| 0 | 0 |
The YES gates in our project consisted of an inducer which triggered the expression from a dedicated promoter. Consequentially, a fluorescent protein and an RNA polymerase are produced. The general scheme of a YES gate is presented in figure 1.
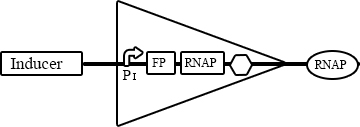
When planing a complex system, there is no way of knowing which combination of promoter and RNAP will work well. Therefore, we planned to attempt the cloning of different RNAPs under the various promoters.
The choice of FPs to follow each promoter was not random. The different considerations will be described below.
YES gate #1
The first YES gate consists of a PTetO promoter which is repressed by TetR. The expression is induced by anhydrotetracycline (aTc). At first, we attempted the cloning of mCitrine downstream to PTetO in plasmids we got from Roee. These showed no fluorescence when tested in the plate reader. Therefore, we turned to the registry for alternatives. We found [http://partsregistry.org/Part:BBa_I13522 BBa_I13522] and [http://partsregistry.org/Part:BBa_I13600 I13600] which express PTetO+GFP and PTetO+CFP respectively.
Continue the tale of failed clones and tell about PTetO+mCherry. Tell about complete YES gate with an RNAP.
YES gate #2
The second YES gate consists of PLac/Ara promoter which is induced by IPTG and arabinose in the presence of LacR and AraC respectively. We got this promoter from Roee followed by the Cerulean gene. This plasmid was used as the starting point for the assembly of YES gate #2. The first step was the addition of a [http://partsregistry.org/Part:BBa_B0015 terminator BioBrick] along with additional restriction sites for the cloning of different RNAPs. However, this cloning step kept failing in several attempts and was abandoned due to lac of time.
YES gate #3
The third YES gate consists of the PLux promoter and a theophylline induced riboswitch (RS). In this YES gate, there is actually a two step regulation. The transcription from the PLux promoter is induced by [http://partsregistry.org/3OC6HSL 3OC6HSL]. The translation is regulated by the RS and is induced by theophylline.
The RS was originally developed in fusion to DsRed Express {1}, which is a red FP from which mCherry was engineered {2}. Since the coding region has effect on the RS function, we thought mCherry would be a good candidate. Unfortunately, this turned out to be a bad choice, since the regulation by the riboswitch failed. The chronicles of the cloning and experimentation with the RS are described in the RS section.
Since the RS regulates the translation of a protein that is fused downstream to it, it had to be fused not only to the FP but also to the RNAPs. This step was completed successfully for several RNAPs as described in the RS section.
References
1. Lynch S. A., Gallivan J. P. 2009. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Research 37(1): 184-192.
2. Shaner N. C., et al. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22(12): 1567-1572.
 "
"