Team:ETH Zurich/PABA/Results
From 2012.igem.org
Contents |
PABA generator
Cloning
We were succesful in constructing both parts <partinfo>BBa_K909014</partinfo> (2699bp) and <partinfo>BBa_K909015</partinfo> (2656bp) based on the already existing parts <partinfo>BBa_K137055</partinfo> and <partinfo>BBa_S04039</partinfo>. The vector backbone is <partinfo>pSB1C3</partinfo> (2079bp). They differ from each other due to the fact that <partinfo>BBa_K909014</partinfo> contains the constitutive promoter <partinfo>BBa_J23100</partinfo> with two NheI restriction sites.
To verify the sizes of the constructs, the plasmids containing <partinfo>BBa_K909014</partinfo> and <partinfo>BBa_K909015</partinfo> were digested with
1. NheI & PstI
- Expected bands of <partinfo>BBa_K909014</partinfo>: 2685 bp & 2061 bp
- Expected bands of <partinfo>BBa_K909015</partinfo>: 4724 bp
2. XbaI & PstI
- Expected bands of <partinfo>BBa_K909014</partinfo>: 2721 bp & 2048 bp
- Expected bands of <partinfo>BBa_K909015</partinfo>: 2678 bp & 2048 bp
As it is visible in Fig.3. the band sizes of the <partinfo>BBa_K909014</partinfo> & <partinfo>BBa_K909015</partinfo> digestion match the expectation.
In a further step <partinfo>BBa_K909015</partinfo> is joined to the PL hybrid promoter <partinfo>BBa_K909011</partinfo> for implementation.
Enzyme overexpression
We were able to overexpress the enzymes pabA, pabB and pabC with our construct. We showed this with an SDS-gel compared to a WT sample. We clearly see a band of the size 22 kDa (pabA) and 71 kDa (pabB/C) which is bigger compared to the WT.
PABA overproduction
We used high-performance liquid chromatography to determine the amount of PABA in our cells. We performed a standard curve with pure PABA. PABA was dissolved as well as the samples in acteonitril and applied to the HPLC.
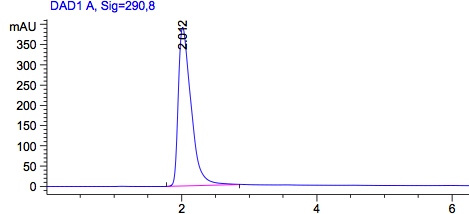
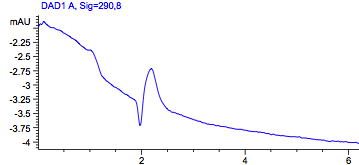
Figure1: HPLC output (290nm) Left: wildtype in acetonitril; Right: Acetonitril without a sample.
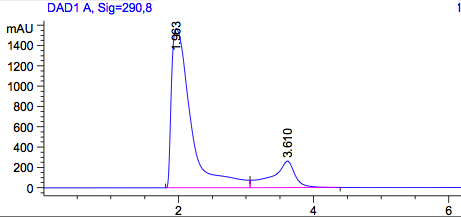
Figure2: HPLC output (290nm) Left: Standard dissolved in acetonitril; Right: Construct in acetonitril.
As a control we used the wildtype Top10. Due to the fact that all bacteria produce PABA as an intermediate of their general biosynthesis, we also expect a signal in this sample. In Figure 1 we see a peak with a retention time of approximately 2 minutes. To be sure that this is the peak of PABA we also used pure PABA dissolved in acetonitril (Figure2), which also shows a retention time of 2 minutes. This graph also shows a second peak, which we could explain with the dissolvent. Dissolved in water we get a a single peak with a retention time of approximately 4 minutes.
To rule out the possibility that this peak comes from the dissolvent itself, we also run acteonitril, which we used to lyse our cells. This control showed no peak.
Figure 2 on the right shows our construct, overexpressing pabA, pabB and pabC. We expect a higher amount of PABA in this sample, an accumulation of PABA. The amount of PABA matches the same concentration as the wildtype. This suggests that our construct could not accumulate PABA in the cell.
PABA is not accumulated in the cell, maybe due to insufficient amount of chorismate in the cell. Therefore we are currently working with an E.coli K12 chorismate overproducer strain [Kast1996], which accumulates chorismate in the cell due to the deficiency in two chorismate mutases. Since chorismate is a precursor molecule we hope to increase PABA output.
References
- Brown, B. a, Headland, L. R., & Jenkins, G. I. (2009). UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochemistry and photobiology, 85(5), 1147–55.
- Christie, J. M., Salomon, M., Nozue, K., Wada, M., & Briggs, W. R. (1999): LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proceedings of the National Academy of Sciences of the United States of America, 96(15), 8779–83.
- Christie, J. M., Arvai, A. S., Baxter, K. J., Heilmann, M., Pratt, A. J., O’Hara, A., Kelly, S. M., et al. (2012). Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science (New York, N.Y.), 335(6075), 1492–6.
- Cloix, C., & Jenkins, G. I. (2008). Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Molecular plant, 1(1), 118–28.
- Cox, R. S., Surette, M. G., & Elowitz, M. B. (2007). Programming gene expression with combinatorial promoters. Molecular systems biology, 3(145), 145. doi:10.1038/msb4100187
- Drepper, T., Eggert, T., Circolone, F., Heck, A., Krauss, U., Guterl, J.-K., Wendorff, M., et al. (2007). Reporter proteins for in vivo fluorescence without oxygen. Nature biotechnology, 25(4), 443–5
- Drepper, T., Krauss, U., & Berstenhorst, S. M. zu. (2011). Lights on and action! Controlling microbial gene expression by light. Applied microbiology, 23–40.
- EuropeanCommission (2006). SCIENTIFIC COMMITTEE ON CONSUMER PRODUCTS SCCP Opinion on Biological effects of ultraviolet radiation relevant to health with particular reference to sunbeds for cosmetic purposes.
- Elvidge, C. D., Keith, D. M., Tuttle, B. T., & Baugh, K. E. (2010). Spectral identification of lighting type and character. Sensors (Basel, Switzerland), 10(4), 3961–88.
- GarciaOjalvo, J., Elowitz, M. B., & Strogatz, S. H. (2004). Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proceedings of the National Academy of Sciences of the United States of America, 101(30), 10955–60.
- Gao Q, Garcia-Pichel F. (2011). Microbial ultraviolet sunscreens. Nat Rev Microbiol. 9(11):791-802.
- Goosen N, Moolenaar GF. (2008) Repair of UV damage in bacteria. DNA Repair (Amst).7(3):353-79.
- Heijde, M., & Ulm, R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends in plant science, 17(4), 230–7.
- Hirose, Y., Narikawa, R., Katayama, M., & Ikeuchi, M. (2010). Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proceedings of the National Academy of Sciences of the United States of America, 107(19), 8854–9.
- Hirose, Y., Shimada, T., Narikawa, R., Katayama, M., & Ikeuchi, M. (2008). Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proceedings of the National Academy of Sciences of the United States of America, 105(28), 9528–33.
- Kast, Asif-Ullah & Hilvert (1996) Tetrahedron Lett. 37, 2691 - 2694., Kast, Asif-Ullah, Jiang & Hilvert (1996) Proc. Natl. Acad. Sci. USA 93, 5043 - 5048
- Kiefer, J., Ebel, N., Schlücker, E., & Leipertz, A. (2010). Characterization of Escherichia coli suspensions using UV/Vis/NIR absorption spectroscopy. Analytical Methods, 9660. doi:10.1039/b9ay00185a
- Kinkhabwala, A., & Guet, C. C. (2008). Uncovering cis regulatory codes using synthetic promoter shuffling. PloS one, 3(4), e2030.
- Krebs in Deutschland 2005/2006. Häufigkeiten und Trends. 7. Auflage, 2010, Robert Koch-Institut (Hrsg) und die Gesellschaft der epidemiologischen Krebsregister in Deutschland e. V. (Hrsg). Berlin.
- Lamparter, T., Michael, N., Mittmann, F., & Esteban, B. (2002). Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proceedings of the National Academy of Sciences of the United States of America, 99(18), 11628–33.
- Levskaya, A. et al (2005). Engineering Escherichia coli to see light. Nature, 438(7067), 442.
- Mancinelli, A. (1986). Comparison of spectral properties of phytochromes from different preparations. Plant physiology, 82(4), 956–61.
- Nakasone, Y., Ono, T., Ishii, A., Masuda, S., & Terazima, M. (2007). Transient dimerization and conformational change of a BLUF protein: YcgF. Journal of the American Chemical Society, 129(22), 7028–35.
- Orth, P., & Schnappinger, D. (2000). Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nature structural biology, 215–219.
- Parkin, D.M., et al., Global cancer statistics, 2002. CA: a cancer journal for clinicians, 2005. 55(2): p. 74-108.
- Rajagopal, S., Key, J. M., Purcell, E. B., Boerema, D. J., & Moffat, K. (2004). Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochemistry and photobiology, 80(3), 542–7.
- Rizzini, L., Favory, J.-J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., Baumeister, R., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science (New York, N.Y.), 332(6025), 103–6.
- Roux, B., & Walsh, C. T. (1992). p-aminobenzoate synthesis in Escherichia coli: kinetic and mechanistic characterization of the amidotransferase PabA. Biochemistry, 31(30), 6904–10.
- Strickland, D. (2008). Light-activated DNA binding in a designed allosteric protein. Proceedings of the National Academy of Sciences of the United States of America, 105(31), 10709–10714.
- Sinha RP, Häder DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci. (2002). 1(4):225-36
- Sambandan DR, Ratner D. (2011). Sunscreens: an overview and update. J Am Acad Dermatol. 2011 Apr;64(4):748-58.
- Tabor, J. J., Levskaya, A., & Voigt, C. A. (2011). Multichromatic Control of Gene Expression in Escherichia coli. Journal of Molecular Biology, 405(2), 315–324.
- Thibodeaux, G., & Cowmeadow, R. (2009). A tetracycline repressor-based mammalian two-hybrid system to detect protein–protein interactions in vivo. Analytical biochemistry, 386(1), 129–131.
- Tschowri, N., & Busse, S. (2009). The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes & development, 522–534.
- Tschowri, N., Lindenberg, S., & Hengge, R. (2012). Molecular function and potential evolution of the biofilm-modulating blue light-signalling pathway of Escherichia coli. Molecular microbiology.
- Tyagi, A. (2009). Photodynamics of a flavin based blue-light regulated phosphodiesterase protein and its photoreceptor BLUF domain.
- Vainio, H. & Bianchini, F. (2001). IARC Handbooks of Cancer Prevention: Volume 5: Sunscreens. Oxford University Press, USA
- Quinlivan, Eoin P & Roje, Sanja & Basset, Gilles & Shachar-Hill, Yair & Gregory, Jesse F & Hanson, Andrew D. (2003). The folate precursor p-aminobenzoate is reversibly converted to its glucose ester in the plant cytosol. The Journal of biological chemistry, 278.
- van Thor, J. J., Borucki, B., Crielaard, W., Otto, H., Lamparter, T., Hughes, J., Hellingwerf, K. J., et al. (2001). Light-induced proton release and proton uptake reactions in the cyanobacterial phytochrome Cph1. Biochemistry, 40(38), 11460–71.
- Wegkamp A, van Oorschot W, de Vos WM, Smid EJ. (2007 )Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl Environ Microbiol. Apr;73(8):2673-81.
 "
"