Team:XMU-China/timedelay
From 2012.igem.org
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Team:XMU-China/css}} | + | {{:Team:XMU-China/css}} |
| - | {{Team:XMU-China/hidden}} | + | {{:Team:XMU-China/hidden}} |
| - | {{Team:XMU-China/titlebar}} | + | {{:Team:XMU-China/titlebar}} |
| - | {{Team:XMU-China/timedelayindex}} | + | {{:Team:XMU-China/timedelayindex}} |
<html xmlns="http://www.w3.org/1999/xhtml"> | <html xmlns="http://www.w3.org/1999/xhtml"> | ||
<head> | <head> | ||
| Line 14: | Line 14: | ||
<body> | <body> | ||
<div id="subpage"> | <div id="subpage"> | ||
| - | |||
<p align="center" class="tit">Time Delay</p> | <p align="center" class="tit">Time Delay</p> | ||
<span class="subtitle"><a name="_Toc01" id="Toc01"></a>1. Overview</span> | <span class="subtitle"><a name="_Toc01" id="Toc01"></a>1. Overview</span> | ||
| - | <p>For the lengths of our genetic circuits are not the same, based on quorum sensing system we had studied last year, we considered that the time of genetic expressions of each circuit is different because of the different lengths and strength of ribosome binding sites. So we constructed circuits with three grades strength of ribosome binding sites, including 0.01, 0.6 and 1.0. After induced by arabinose, the duration of response time for GFP expression | + | <p>For the lengths of our genetic circuits are not the same, based on quorum sensing system <sup><a href="#_ENREF_1" title="Ron Weiss, 2003 #2">[1]</a></sup>we had studied last year, we considered that the time of genetic expressions of each circuit is different because of the different lengths and strength of ribosome binding sites. So we constructed circuits with three grades strength of ribosome binding sites, including 0.01, 0.6 and 1.0. After induced by arabinose, the duration of response time for GFP expression increases as the strength of RBS declines, bringing about a time-course display.</p><hr> |
<p> </p> | <p> </p> | ||
<span class="subtitle"><a name="_Toc02" id="Toc02"></a>2. Review of last year</span> | <span class="subtitle"><a name="_Toc02" id="Toc02"></a>2. Review of last year</span> | ||
| - | <p>Last year, we had developed a series of devices which program a bacteria population to maintain at different cell densities. [ | + | <p>Last year, we had developed a series of devices which program a bacteria population to maintain at different cell densities. <sup><a href="#_ENREF_2" title="Ron Weiss, 2003 #2">[2]</a></sup>We had designed and characterized the genetic circuit to establish a bacterial ‘population-control’ device in <em>E. coli</em> based on the well-known quorum-sensing system from <em>Vibrio fischeri</em>, which autonomously regulates the density of an <i>E. coli</i> population. The cell density however was influenced by the expression levels of a killer gene (<i>ccdB</i>) in our device. As such, we had successfully controlled the expression levels of CcdB by using RBSes of different strength and mutated luxR promoters (<em>lux pR</em>). This circuit incorporated a mechanism for programmed death in respond to changes in the environment. <br /> |
</p><p align="center"> <img src="https://static.igem.org/mediawiki/2012/1/16/Timedelay_f1.png" alt="" width="508" height="349" /><br /> | </p><p align="center"> <img src="https://static.igem.org/mediawiki/2012/1/16/Timedelay_f1.png" alt="" width="508" height="349" /><br /> | ||
| - | Figure 1 | + | <b>Figure 1:</b> The mechanism graph of iccdB last year<sup><a href="#_ENREF_3" title="Ron Weiss, 2003 #2">[3]</a></sup></p><hr><br> |
| - | + | ||
| - | + | ||
| - | + | ||
<span class="subtitle"><a name="_Toc03" id="Toc03"></a>3. Constructions this year</span> | <span class="subtitle"><a name="_Toc03" id="Toc03"></a>3. Constructions this year</span> | ||
| - | <p align="center"><img src="https://static.igem.org/mediawiki/2012/ | + | <p align="center"><img src="https://static.igem.org/mediawiki/2012/e/e6/Delay_01.jpg" alt="" width="420" /><br /> |
| - | Figure 2 | + | <b>Figure 2:</b> The construction devices of time delay system<br /> |
| - | <p>Based on our last year’s project, for convenience of testing the results of time delay device, we chose GFP instead of | + | <p align="left">Based on our last year’s project, for convenience of testing the results of time delay device, we chose GFP instead of CcdB as our reporter. On the other hand, because of the basal expression of <em>P<sub>lac</sub></em> is very strong, we decided to use arabinose to induce <em>P<sub>BAD</sub></em> promoter to activate our device.<sup><a href="#_ENREF_4" title="Ron Weiss, 2003 #2">[4]</a></sup> Meanwhile, to accomplish the aim of time delay, which means we should make the GFP produce in different period of time, we decided to change the RBS’s strength of <i>luxI</i> expression system. RBSes of different strengths will cause the speed of quorum sensing effect, which will lead to the delay of GFP production time.<sup><a href="#_ENREF_5" title="Ron Weiss, 2003 #2">[5]</a><a href="#_ENREF_6" title="Ron Weiss, 2003 #2">[6]</a></sup></p><br /> |
| - | <img src="https://static.igem.org/mediawiki/2012/1/10/Timedelay_f3.png" alt="" width="428" height="241" /><br /> | + | <p align="center"><img src="https://static.igem.org/mediawiki/2012/1/10/Timedelay_f3.png" alt="" width="428" height="241" /><br /> |
| - | Figure 3 | + | <b>Figure 3:</b> The mechanism graph of time delay system</p><hr><br> |
| + | |||
<span class="subtitle"><a name="_Toc04" id="Toc04"></a>4. Results and discussion | <span class="subtitle"><a name="_Toc04" id="Toc04"></a>4. Results and discussion | ||
| - | |||
</span> | </span> | ||
<p><span class="subsubtitle">4.1 Optimization of inducer concentration</span><br /> | <p><span class="subsubtitle">4.1 Optimization of inducer concentration</span><br /> | ||
| - | + | <p align="center" > <img src="https://static.igem.org/mediawiki/2012/0/0f/TimedelayFig4.png" alt="" width="491" height="347" /><br> | |
| - | + | <b>Figure 4:</b> The optimization of the arabinose concentration added into the TD1.0 including<br> | |
| + | 0, 0.1, 1.0, 10 mM arabinose.<a target="_blank" href=" https://static.igem.org/mediawiki/2012/e/e4/-Fig_4-Optimization_of_arabinose_induced_concentration_%28TD1.0%29.jpg">[click to data]</a></p><br> | ||
<p align="left">We can find that the curve with 0.1 mM arabinose has a higher fluorescence. So we chose the </a>0.1 mM arabinose for the following fluorescence tests.</p> | <p align="left">We can find that the curve with 0.1 mM arabinose has a higher fluorescence. So we chose the </a>0.1 mM arabinose for the following fluorescence tests.</p> | ||
<p class="subsubtitle">4.2 Fluorescent test of the time delay devices</p> | <p class="subsubtitle">4.2 Fluorescent test of the time delay devices</p> | ||
<br clear="all" /> | <br clear="all" /> | ||
<div> | <div> | ||
| - | <p align="center"><img src="https://static.igem.org/mediawiki/2012/ | + | <p align="center"><img src="https://static.igem.org/mediawiki/2012/8/8c/XMUtimedelay5_6_7.png" alt="" width="500" /><br /> |
| - | Figure 5 | + | <b>Figure 5:</b> Fluorescence curves of strain TD1.0, TD0.6 and TD0.01 <br> |
| - | + | induced | |
| - | + | with 0.1 mM arabinose and without arabinose.<a target="_blank" href=" https://static.igem.org/mediawiki/2012/6/6a/XMUtimedelay5_6_7data.png">[click to data]</a><br /> | |
| + | </p> | ||
</div> | </div> | ||
<br clear="all" /> | <br clear="all" /> | ||
<div> | <div> | ||
| - | <p align="center"><img src="https://static.igem.org/mediawiki/2012/ | + | <p align="center"><img src="https://static.igem.org/mediawiki/2012/f/fc/Delay_02.jpg" alt="" width="449" height="334" /><br /> |
| - | Figure | + | <b>Figure 6:</b> Fluorescence curves of the induced TD0.01, TD0.6, TD1.0 and <br> |
| - | + | the control BL21(DE3).<a target="_blank" href=" https://static.igem.org/mediawiki/2012/6/6f/-Fig_8-Comparison_of_TD0.01_0.6_1.0.jpg">[click to data]</a><br /> | |
| - | + | ||
<img src="https://static.igem.org/mediawiki/2012/0/00/TimedelayFig_9.png" alt="" width="480" height="371" /><br /> | <img src="https://static.igem.org/mediawiki/2012/0/00/TimedelayFig_9.png" alt="" width="480" height="371" /><br /> | ||
| - | Figure | + | <b>Figure 7:</b>The separate time taken up for the three circuits' fluorescent value to reach 5000 <br> |
| - | + | after induction, which shows that the stronger RBS circuit contains, the less time it takes.<br> | |
| - | <p>Compared with the groups absence of arabinose, the fluorescence of | + | </p> |
| - | </div><hr> | + | <p>Compared with the groups absence of arabinose, the fluorescence of induced groups were much higher. On the other hand, we could find even the circuit with RBS<sub>0.01</sub>, the most insensitive one, also expressed green fluorescence protein without arabinose, which means basal expressing phenomenon. According to figure 5, when the three cultures induced by the same concentration of arabinose, the fluorescence of TD1.0 was the highest, TD0.6 was middle and TD0.01 was the lowest. Finally, as shown in figure 7, the fluorescence of the circuits with different RBSes also reached a certain level in different time. It fit the goal of time delay.</p> |
| + | </div><hr><br> | ||
<p class="subtitle"><a name="_Toc05" id="Toc05"></a>5. Reference | <p class="subtitle"><a name="_Toc05" id="Toc05"></a>5. Reference | ||
</p> | </p> | ||
| - | <a | + | <a name="_ENREF_1" id="_ENREF_1">[1]W.Claiborne Fuqua, S.C.W., E. Peter Greenberg, Quorum Sensing in Bacteria: the LuxR-LuxI Family of Cell Density-Responsive Transcriptional Regulatorst. JOURNAL OF BACTERIOLOGY, 1994. 176(2): p. 269-275.<br> |
| - | + | </a> | |
| + | <a name="_ENREF_2" id="_ENREF_2">[2]You, L., et al., Programmed population control by cell-cell communication and regulated killing. Nature, 2004. 428(6985): p. 868-71.</a> | ||
| + | <br> | ||
| + | <a name="_ENREF_3" id="_ENREF_3">[3]https://2011.igem.org/Team:XMU-China.<br> | ||
| + | </a> | ||
| + | <a name="_ENREF_4" id="_ENREF_4">[4]Mangan, S., et al., The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol, 2006. 356(5): p. 1073-81. | ||
| + | </a><br> | ||
| + | <a name="_ENREF_5" id="_ENREF_5"> | ||
| + | [5]Camas, F.M., J. Blazquez, and J.F. Poyatos, Autogenous and nonautogenous control of response in a genetic network. Proc Natl Acad Sci U S A, 2006. 103(34): p. 12718-23. | ||
| + | </a><br> | ||
| + | <a name="_ENREF_6" id="_ENREF_6">[6]Alon, U., Network motifs: theory and experimental approaches. Nat Rev Genet, 2007. 8(6): p. 450-61. | ||
| + | </a> | ||
</div> | </div> | ||
| - | + | ||
| - | + | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 02:57, 27 October 2012
 |  |
 |  |
| Contents[hide][show] |
|---|
Time Delay
1. OverviewFor the lengths of our genetic circuits are not the same, based on quorum sensing system [1]we had studied last year, we considered that the time of genetic expressions of each circuit is different because of the different lengths and strength of ribosome binding sites. So we constructed circuits with three grades strength of ribosome binding sites, including 0.01, 0.6 and 1.0. After induced by arabinose, the duration of response time for GFP expression increases as the strength of RBS declines, bringing about a time-course display.
2. Review of last year
Last year, we had developed a series of devices which program a bacteria population to maintain at different cell densities. [2]We had designed and characterized the genetic circuit to establish a bacterial ‘population-control’ device in E. coli based on the well-known quorum-sensing system from Vibrio fischeri, which autonomously regulates the density of an E. coli population. The cell density however was influenced by the expression levels of a killer gene (ccdB) in our device. As such, we had successfully controlled the expression levels of CcdB by using RBSes of different strength and mutated luxR promoters (lux pR). This circuit incorporated a mechanism for programmed death in respond to changes in the environment.
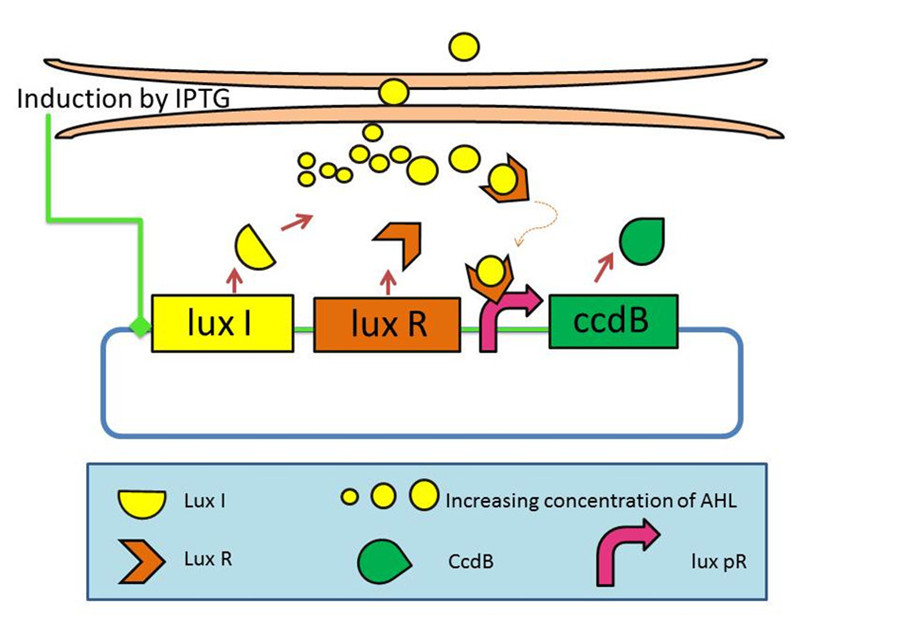
Figure 1: The mechanism graph of iccdB last year[3]
3. Constructions this year

Figure 2: The construction devices of time delay system
Based on our last year’s project, for convenience of testing the results of time delay device, we chose GFP instead of CcdB as our reporter. On the other hand, because of the basal expression of Plac is very strong, we decided to use arabinose to induce PBAD promoter to activate our device.[4] Meanwhile, to accomplish the aim of time delay, which means we should make the GFP produce in different period of time, we decided to change the RBS’s strength of luxI expression system. RBSes of different strengths will cause the speed of quorum sensing effect, which will lead to the delay of GFP production time.[5][6]

Figure 3: The mechanism graph of time delay system
4. Results and discussion
4.1 Optimization of inducer concentration

Figure 4: The optimization of the arabinose concentration added into the TD1.0 including
0, 0.1, 1.0, 10 mM arabinose.[click to data]
We can find that the curve with 0.1 mM arabinose has a higher fluorescence. So we chose the 0.1 mM arabinose for the following fluorescence tests.
4.2 Fluorescent test of the time delay devices

Figure 5: Fluorescence curves of strain TD1.0, TD0.6 and TD0.01
induced
with 0.1 mM arabinose and without arabinose.[click to data]

Figure 6: Fluorescence curves of the induced TD0.01, TD0.6, TD1.0 and
the control BL21(DE3).[click to data]

Figure 7:The separate time taken up for the three circuits' fluorescent value to reach 5000
after induction, which shows that the stronger RBS circuit contains, the less time it takes.
Compared with the groups absence of arabinose, the fluorescence of induced groups were much higher. On the other hand, we could find even the circuit with RBS0.01, the most insensitive one, also expressed green fluorescence protein without arabinose, which means basal expressing phenomenon. According to figure 5, when the three cultures induced by the same concentration of arabinose, the fluorescence of TD1.0 was the highest, TD0.6 was middle and TD0.01 was the lowest. Finally, as shown in figure 7, the fluorescence of the circuits with different RBSes also reached a certain level in different time. It fit the goal of time delay.
[1]W.Claiborne Fuqua, S.C.W., E. Peter Greenberg, Quorum Sensing in Bacteria: the LuxR-LuxI Family of Cell Density-Responsive Transcriptional Regulatorst. JOURNAL OF BACTERIOLOGY, 1994. 176(2): p. 269-275.
[2]You, L., et al., Programmed population control by cell-cell communication and regulated killing. Nature, 2004. 428(6985): p. 868-71.
[3]https://2011.igem.org/Team:XMU-China.
[4]Mangan, S., et al., The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol, 2006. 356(5): p. 1073-81.
[5]Camas, F.M., J. Blazquez, and J.F. Poyatos, Autogenous and nonautogenous control of response in a genetic network. Proc Natl Acad Sci U S A, 2006. 103(34): p. 12718-23.
[6]Alon, U., Network motifs: theory and experimental approaches. Nat Rev Genet, 2007. 8(6): p. 450-61.
 "
"