Team:Valencia Biocampus/Molecular
From 2012.igem.org

Molecular mechanisms
Bacteria
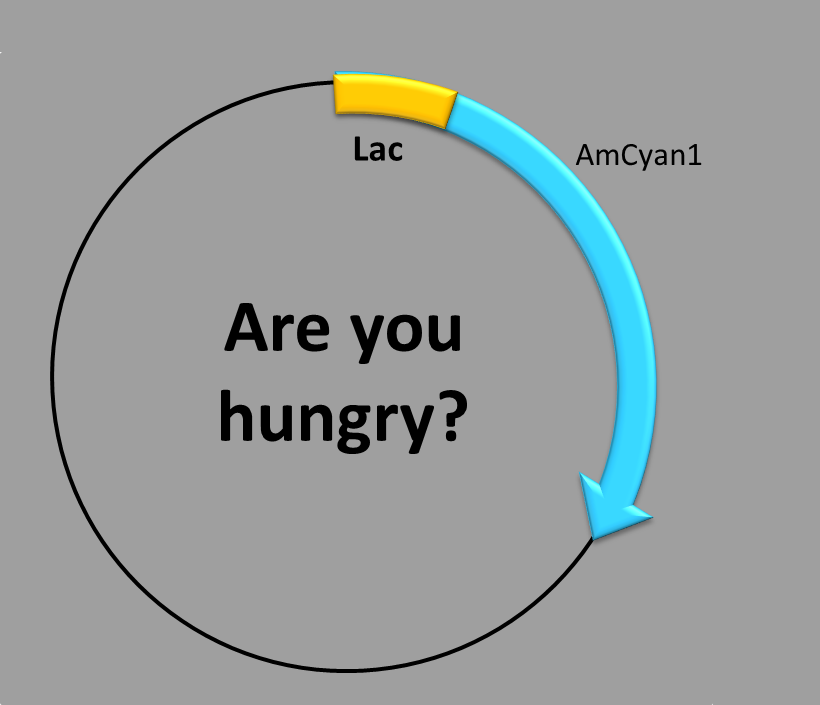
LACTOSE-INDUCED PROMOTER
This construction is made up of three parts:
- The transcription factor-binding site inside the promoter,
- the repressor-binding site outside the promoter and
- the coding sequence, which contains a synthetic fluorescent (blue) protein. Our construction uses the well-known lactose operon system [1]. Since there is an operator region that blocks transcription, it is necessary to know it and avoid it.
When is the protein synthesized? In order to obtain the blue fluorescent protein two conditions have to be met.
First condition: there is no glucose in the medium. Second condition: lactose is present in the medium (it also works with other inductors, like IPTG).
The molecular mechanism underlying this phenomenon is as follows: a lack of glucose promotes the formation of CAP (or CRP [2]), which binds to specific sites upstream of sugar-metabolizing genes and activates its transcription. The binding of this molecule depends on the presence of the allosteric effector cAMP
(the concentration of this metabolite changes in response to the presence or absence of different nutrients). Moreover, another condition is needed since LacI repressor, produced constitutively by the lacI gene inside the lactose operon, will bind to the operator region and will block the transcription unless lactose is also present in the medium. Once lactose enters the cell it is converted to allolactose [3], and this molecule binds tightly to the repressor so it can no longer block the transcription. Then, the fluorescent protein can glow in the cytoplasm!
How did we deal with this construction? In our experiments we not only tested the differential expression when glucose is absent + lactose present and vice versa, but we also tested the expression and growth rates when different compounds were added as carbon-enriched sources. For example, we added sodium acetate and galactose as substitutes of glucose. We determined the best IPTG concentration for our cultures too.
References:
- F. Jacob and J. Monod. (1959) Genes of structure and genes of regulation in the biosynthesis of proteins. C. R. Hebd. Seances Acad. Sci.249: 1282–4.
- S., Busby and R.H., Ebright. (2001). Transcription activation by catabolite activator protein (CAP). J. Mol. Biol.293: 199–213.
- RE., Huber, K., Wallenfels and G.,Kurz. (1975)The action of beta-galactosidase (Escherichia coli) on allolactose.Canadian Journal of Biochemistry, 53(9):1035-1038

*For lactose-activation, switch glucose for IPTG.
Glucose concentration in lactose-activation media (from a stock of 50 g/L): 5 g/L, 1 g/L and 0.1 g/L. Since 0.1 g/L is likely to be the most useful concentration, let’s go on later with 0.05 g/L, 0.01 g/L and 0.001 g/L. Remember to add IPTG!
Check other molecular mechanisms or see the characterisation of this construction.
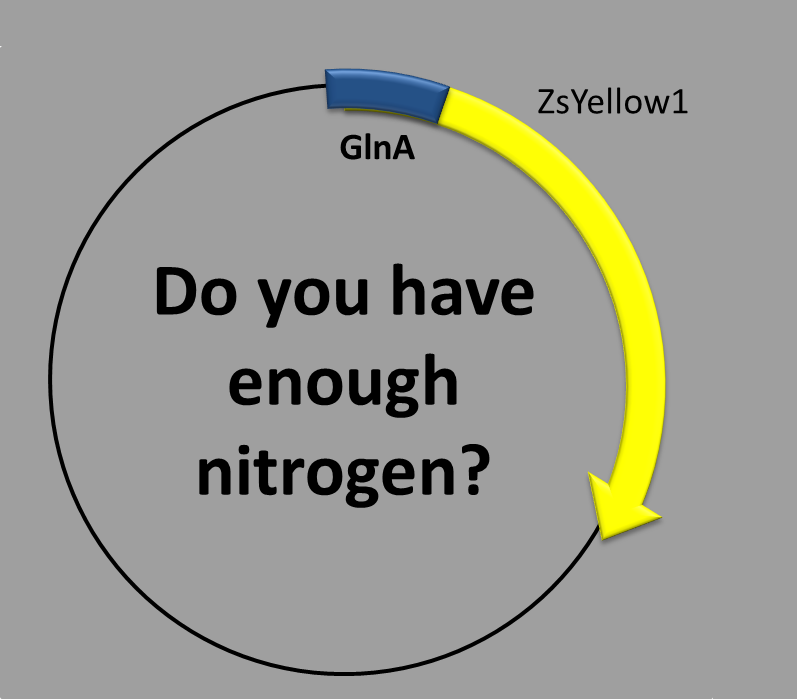
NITROGEN-REGULATED PROMOTER
This construction contains two parts:
- The transcription factor-binding site inside the promoter and
- the coding sequence, which contains a synthetic fluorescent (yellow) protein. We chose as the promoter sequence the one of the glnA gene. There is long evidence that this promoter is regulated by nitrogen concentration [1]. Moreover, the promoter is not the canonical one (sigma70) but an alternative one (sigma54) [2,3].
When is the protein synthesized? In order to obtain the yellow fluorescent protein a condition should to be met. That condition is related to nitrogen-starvation. The less nitrogen there is, the more expression you get!
The molecular mechanism underlying this phenomenon is as follows: in gram-negative bacteria, transcriptional activation in response to some external stimuli (absence of nitrogen, high UV-dosis, for example) often involves the alternative sigma factor, sigma54. This factor, also called RpoN or sigma N, was originally identified as the sigma factor for nitrogen-controlled genes. sigma54 works in conjunction with members of the NtrC (Nitrogenregulatoryprotein C) superfamily of transcriptional activators. In our case, when ammonia levels are low the bacterium undergoes some metabolic changes. Within these changes, there are some related to nitrogen assimilation and processing, so our construction, which responds to low ammonia levels, increases its transcription.
How did we deal with this construction?We developed a nitrogen-gradation tube experiment, which means that we added different (NH4)2SO4 concentrations to each tube, so we can measure a gradation of the protein fluorescence. We used a synthetic medium for this.
References:
- Reitzer, L.J., Movsas, B. andMagasanik, B. (1989)Activation of glnA transcription by Nitrogen Regulator I (NRI)- phosphate in Escherichia coli: evidence for a long-range physical interaction between NRI-phosphate and RNA polymerase.Journal of bacteriology, 171 (10):5512-5522
- Wang, L., Guo, Y. and Gralla, J.D. (1999) Regulation of sigma 54-dependent transcription by core promoter sequences: role of - 12 region nucleotides. Journal of bacteriology, 181 (24):7558–7565
- Barrios, H., Valderrama, B. y Morett, E. (1999) Compilation and analysis of sigma54-dependent promoter sequences. Nucleics Acids Research, 27 (22):4305-4313.

Check other molecular mechanisms or see the characterisation of this construction.
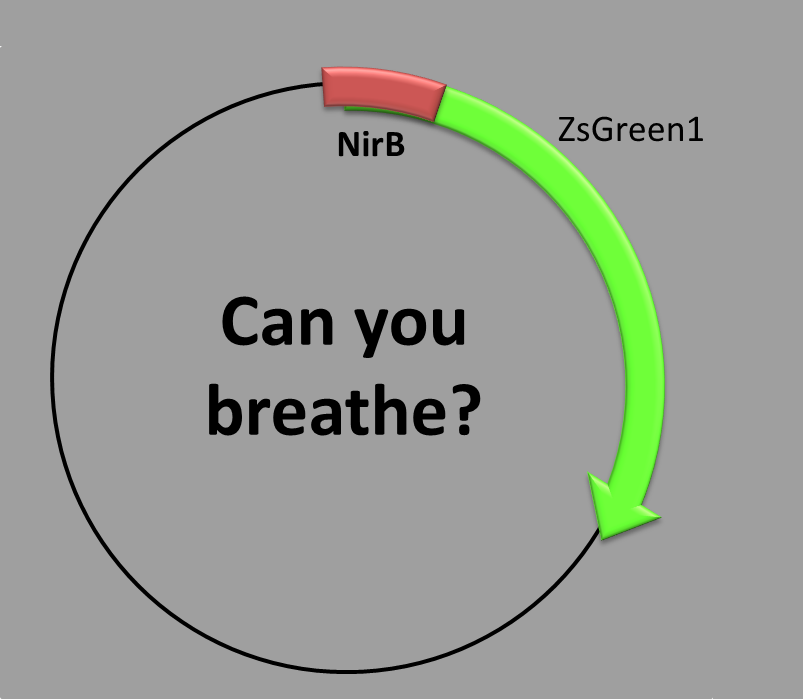
OXYGEN-REGULATED PROMOTER
This construction has two parts:
- The transcription factor-binding site inside the promoter and
- the coding sequence, which contains a synthetic fluorescent (green) protein. We chose as the promoter sequence the one of the nirB gene, because it is 100% dependent on FNR [1,2], which is the transcription factor activated when there is no oxygen in the environment. It is involved mainly in cellular respiration and carbon metabolism during anaerobic cell growth.
When is the protein synthesized? In order to obtain this green fluorescent protein one condition is required: anaerobic conditions should be met. We grew the bacteria in a oxygen-controlled environment [3].
The molecular mechanism underlying this phenomenon is as follows: the promoter is activated when there is a lack of O2 in the environment thanks to a family of proteins called FNR. FNR proteins are global transcriptional regulators that respond to fluctuations in ambient oxygen. They recognize DNA sequences in inverted disposition with this sequence: TTGATN1N2N3N4ATCAA [4], where N1-4 represents a non-conserved tetrad (no influence). The FNR sensor domain contains four cysteine residues that act like ligands for a group (cluster) of 4Fe-4S oxygen-sensitive structures. The acquisition of a 4Fe-4S cluster begins heterodimer formation of FNR that bind to specific sites of DNA and regulate transcription of target promoters. Under aerobic conditions (i.e. when oxygen is present), 4Fe-4S clusters are disassembled and the FNR dimers dissociate and form no structures that can bind to DNA.
How did we deal with this construction? Anaerobic conditions were achieved after putting a culture of E. coli, stirring, inside a sealed box. We let the bacteria to grow for at least two days and then we measured fluorescence.
References:
- P.S., Jayaraman, K.L., Gaston, J.A., Cole and S. Busby. (1988) The nirB promoter of Escherichia coli: location of nucleotide sequences essential for regulation by oxygen, the FNR protein and nitrite. Mol Microbiol. 2(4):527-30.
- D.F., Browning, J.A. Cole and S. Busby. (2008) Regulation by nucleoid-associated proteins at the Escherichia coli nir operon promoter. J Bacteriol. 190(21):7258-67.
- S., Becker, G., Holighaus, T., Gabrielczyk and G., Unden. (1996) O2 as the Regulatory Signal for FNR-Dependent Gene Regulation in Escherichia coli. Journal of Bacteriology, (178): 4515–4521.
- C., Scott, J.D., Partridge, J.R. Stephenson and J. Green. (2003) DNA target sequence and FNR-dependent gene expression. FEBS Letters 541: 97-101.

*Upper oil layer for anaerobiosis
Check other molecular mechanisms or see the characterisation of this construction.
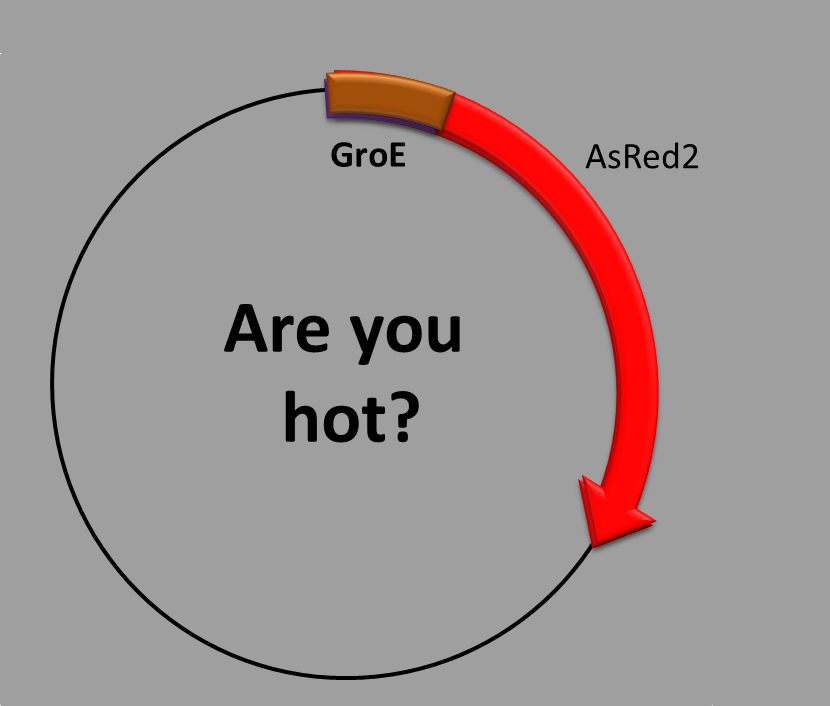
TEMPERATURE-INDUCED PROMOTER
This construction is made up of three parts:
- The transcription factor-binding site inside the promoter
- the coding sequence, which contains a synthetic fluorescent (red) protein. Our construction uses the well-known high chaperon expression when the bacterium
is suffering from heat shock [1]. We have taken the promoter sequence of groE, one of these proteins highly produced when heat shock [2].
When is the protein synthesized? In order to obtain the red fluorescent protein a condition has to be met. High temperatures for E. coli must be reached, like for example from 42ºC degrees. We focused on 44ºC because here we got the best results.
The molecular mechanism underlying this phenomenon is as follows: high temperatures are sometimes harmful for microorganisms, so they have adapted successfully for such a strong condition by synthesizing some proteins, called HSP (Heat Shock Proteins), that help in protein folding. Heat-shock response is mediated by the sigma32 factor [3]. This alternative sigma factor allows the RNA polymerase to bind to some consensus promoter sequence. groE has this sequence in its promoter [4]. That is because this gene encodes a chaperon protein, GroE. Chaperone proteins are a group of proteins present in all cells; many of them are heat shock proteins, whose function is to assist the folding of other proteins in the newly formed protein synthesis.
In the case of GroE, it processes a nonnative polypeptide in a cycle consisting of three steps. First, the polypeptide substrate is captured by GroEL. Upon binding of the co-chaperone GroES and ATP, the substrate is then discharged into a unique microenvironment inside of the chaperone, which promotes proper folding. After hydrolysis of ATP, the polypeptide is released into solution. Moreover, GroE may actively increase the folding efficiency, e.g. by unfolding of misfolded protein molecules. This chaperon has an important role in heat shock process too, helping other proteins not to denature.
How did we deal with this construction? We were able to activate the promoter after a heat shock, like keeping the culture at 44ºC during 5 min. However, since we wanted a very good modeling for this construction, we developed a lot of experiments with different temperatures, different times of incubation, as well as gradating the temperature ranging from 37ºC to 46ºC.
References:- De Maio A (1999) Heat shock proteins: facts, thoughts, and dreams. Shock, 11 (1): 1–12.
- Kusukawa, N., Yura, T., Ueguchi, C., Akiyama, Y. And Ito, K. (1989) Effects of mutations in heat shock genes groES and groEL on protein export in Escherichia coli. EMBO J 8 (11); 3517-21.
- Nonaka, G., Blankschien, M., Herman, C., Gross, C.A. and Rhodius, V.A. (2006) Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, revelas a multifaceted celular response toheat stress. Genes and Development, 20:1776-1789.
- Cowing, D.H., Bardwell, J.C., Craig, E.A., Woolford, C., Hendrix, R.W. and Gross, C.A. (1985) Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci USA, 82: 2679-2683.

*Heat shock at 42ºC (5min)
Check other molecular mechanisms or see the characterisation of this construction.
UV-INDUCED PROMOTER
This construction is made up of two parts:
- The transcription factor-binding site inside the promoter
- The repressor-binding site inside the promoter and
- the coding sequence, which contains a synthetic fluorescent (green) protein. The promoter was taken from the RecA one. Proteins that participate in the orchestrated bacterial SOS-response [1] that involves more than forty independent SOS genes, most of which encode protein sengaged in protection, repairing and replication regulate our construction.
When is the protein synthesized? In order to obtain the green fluorescent protein one condition should be met. Since there is a repressor, lexA, that blocks any possible transcription, it is needed to UV-irradiate the cells to break down lexA and habilitate transcription.
The molecular mechanism underlying this phenomenon is as follows: when a bacterium undergoes severe damage it uses this repair system for maintaining a functional (double stranded) DNA able to duplicate. The damage can be so harmful that it can generate ruptures, dimers, bases modification, etc., therefore the bacteria releases RecA resulting in polymerized and filamentous structures which bind to and stabilize DNA [2], letting some error-prone DNA polymerases (translesion synthesis) to come and copy, randomly, the DNA. Solving the problem this way provokes several mutations in the bacteria's genome but it will stay alive because it maintains a functional DNA. Activation of the polymerases IV and V only occurs in advance stages of the response.The system comprises a negative regulator of the SOS response (LexA) and a positive regulator of the SOS response (RecA). The idea is that UV irradiation promotes LexA autocatalytic activity and RecA is activated. Then, LexA will not be able to bind to the promoters (SOS boxes or LexA boxes) and transcription will start (here is our construction with the protein we want). The process is relatively slow, reaching higher values of expression more than one hour later [3].
In our construction, we have picked out the LexA-binding site inside RecA promoter and then we have inserted a GFP sequence. Then, after irradiating we can see fluorescence… E. coli has accomplished our order!
How did we deal with this construction? We designed a UV-irradiation controlling device called Tordeitor 3000, so we could emit UV (254nm) without risk. After putting the plates, we irradiate different times in order to expose E. coli to different energies [4,5,6].
References:
- Radman, B. (1975) "Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis". Basic Life Sciences 5A: 355–367.
- Michael, B. (2005). "After 30 Years of Study, the Bacterial SOS Response Still Surprises Us". PLoSBiology 3 (7): e255.
- Friedman, N., Vardi, S., Ronen, M., Alon, U. and Stavans J. (2005) Precise Temporal Modulation in the Response of the SOS DNA Repair Network in Individual Bacteria. PLoSBiology 3 (7): e238.
- Cutter, K.L., Alloush, H.M. and Salisbury, V.C. (2007) Stimulation of DNA repair and increased light output in response to UV irradiation in Escherichia coli expressing lux genes. Luminescence 22: 177–181.
- Janion, C. (2008) Inducible SOS Response System of DNA Repair and Mutagenesis in Escherichia coli. Int. J. Biol. Sci. 4.
- Krishna, S., Maslov, S. and Sneppen, K. (2007) UV-Induced Mutagenesis in Escherichia coli SOS Response: A Quantitative Model. PLoSBiology 3 (3): e41.

*UV irradiation with a lamp
Check other molecular mechanisms or see the characterisation of this construction.
Yeast
FERMENTATIVE RESPONSE
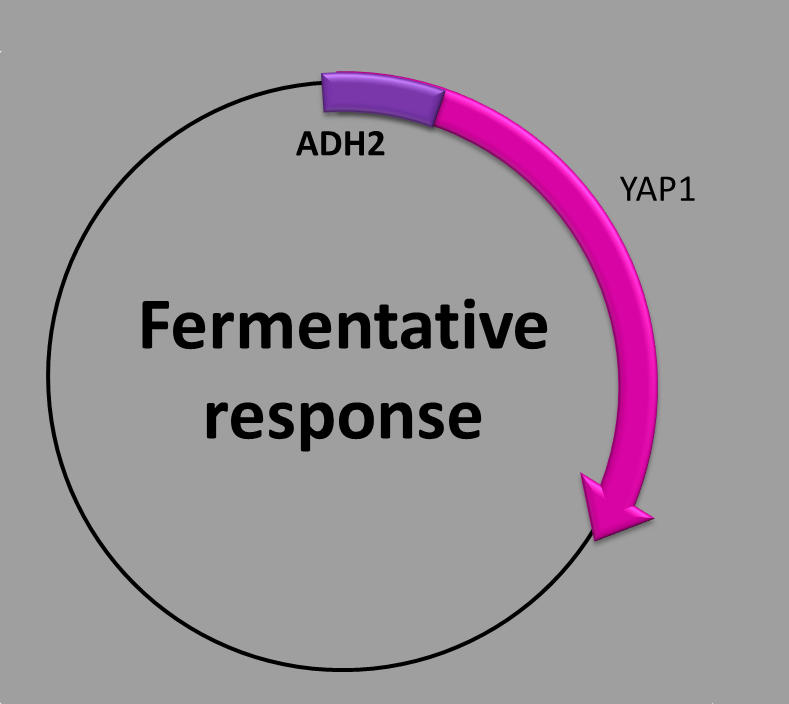
This construction has three parts: (1) a transcription factor binding site inside the promoter (UAS1), (2) a transcription factor binding site (UAS2/CSRE), and (3) the coding sequence which contains the yeast AP-1 protein (YAP1). We used the ADH2 promoter, which is regulated by two trans-acting elements, both necessary for maximal promoter activity in the absence of a fermentable carbon source [1].
When is the protein synthesized? Our construction is repressed several hundred-fold in the presence of glucose, so the transcription of YAP1 protein is initiated once the glucose in the medium is depleted.
The molecular mechanism underlying this phenomenon is as follows: there are two transcription factors which regulate the promoter, ADR1 a carbon source-sensor zinc-finger [2] and CAT8 [3]. In the absence of glucose both cis-acting sites in the promoter are bound cooperatively by the transcriptional activators. ADR1 binds to the UAS1 site while CAT8 binds to the UAS2/CSRE site and regulates positively the transcription of YAP1 protein by the activation of the ADH2 promoter [4]. The presence of glucose downregulates the levels of the transcription factors (ADR1 and CAT8) which results in the depletion of the production of YAP1 protein by the repression of the ADH2 promoter.
- Tachibana, C., Yoo, J.Y., Tagne, J.B., Kacherovsky, N., Lee, T.I. and Young, E.T. (2005) Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8.Mol Cell Biol 25(6):2138-46
- Denis, C.L. and Young, E.T. (1983) Isolation and characterization of the positive regulatory gene ADR1 from Saccharomyces cerevisiae. Mol Cell Biol3(3):360-70.
- Hedges, D., Proft, M. and Entian, K.D. (1995) CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol 15(4):1915-22
- Walther, K. and Schuller, H.J. (2001) Adr1 and Cat8 synergistically activate the glucose-regulated alcohol dehydrogenase gene ADH2 of the yeast Saccharomyces cerevisiae. Microbiology 147(Pt 8):2037-44
GO BACK AND CHECK OTHER YEAST CONSTRUCTIONS
OXIDATIVE STRESS RESPONSE
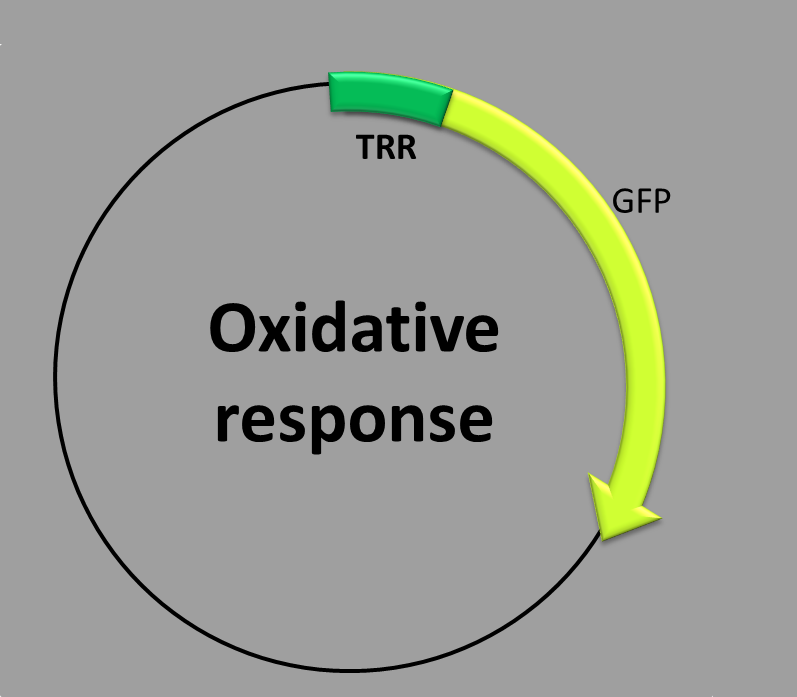
This construction is made up of two parts: (1) a transcriptional factor binding site inside the promoter, (2) the coding sequence which contains a synthetic fluorescent (green) protein. The expression of the thioredoxin reductase (TRR) promoter is regulated by two transcription factors, YAP1 and SKN7, which work together in response to oxidative stress [1].
When is the protein synthesized? We obtain the green fluorescent protein by the addition of hydrogen peroxide. (H2O2).
The molecular mechanism underlying this phenomenon is as follows: YAP1 transcription factor is regulated by subcellular localization. Under normal conditions an exporting receptor (CRM1) escorts YAP1 from the nucleus to the cytoplasm where it is stored. In the presence of hydrogen peroxide, another protein (HYR1) catalyzes the formation of an intramolecular disulfide bond in YAP1 [2]. This conformational change allows YAP1 to accumulate in the nucleus and to bind to the TRR promoter by a bZIP DNA-binding domain. On the other side, SKN7 transcription factor is constitutively nuclear. In the presence of hydrogen peroxide, SKN7 is able to associate with the TRR promoter. Some studies suggest that the formation of the SKN7+ YAP1+ DNA complex requires more than the binding of these two transcription factor to the promoter in close proximity. In conclusion, the two transcriptional factors require only one binding site within the promoter [3]. Once the two proteins are bound to the TRR promoter, the transcription of the green fluorescent protein starts.
- Lee, J., Godon, C., Lagniel, G., Spector, D., Garin, J., Labarre, J. and Toledano, B. (1999) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. The journal of biological chemistry 23(274): 16040-16046.
- Kuge, S., Jones, N. and Nomoto, A. (1997) Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J 16(7):1710-20
- Mulford, K.E. and Fassler, J.S. (2011) Association of the Skn7 and Yap1 transcriptiom factors in the Saccharomyces cerevisiae oxidative Stress response. Eukaryotic cell 10(6):761
GO BACK AND CHECK OTHER YEAST CONSTRUCTIONS - De Maio A (1999) Heat shock proteins: facts, thoughts, and dreams. Shock, 11 (1): 1–12.
 "
"