Team:USP-UNESP-Brazil/Plasmid Plug n Play/Background
From 2012.igem.org
Andresochoa (Talk | contribs) |
Andresochoa (Talk | contribs) |
||
| Line 20: | Line 20: | ||
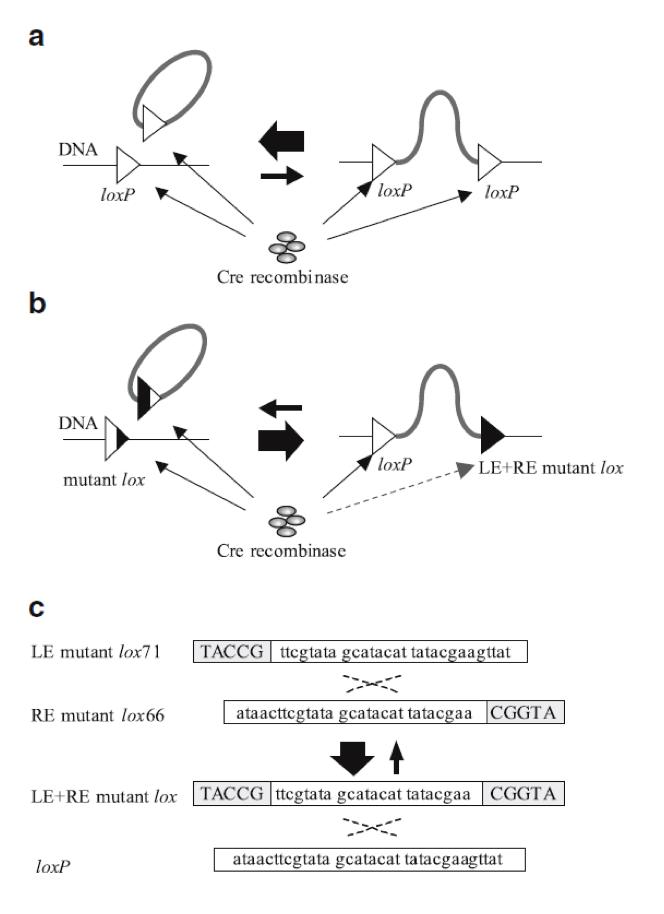
| - | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=fig2plugplay.JPG | caption= Fig 2. Illustration of Cre-loxP and Cre-LE and RE-mutant lox system. Black regions of the triangles represent sites at which nucleotide sequence changes occurred. A) Recombination between loxP sites. B) Recombination between a LE mutant lox71 and a RE mutant lox66 produces a loxP and a LE + RE mutant lox. C) Gray boxes indicate sites at which nucleotide sequence changes occurred (6). | size=600px }} | + | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=fig2plugplay.JPG | caption= Fig. 2. Illustration of Cre-loxP and Cre-LE and RE-mutant lox system. Black regions of the triangles represent sites at which nucleotide sequence changes occurred. A) Recombination between loxP sites. B) Recombination between a LE mutant lox71 and a RE mutant lox66 produces a loxP and a LE + RE mutant lox. C) Gray boxes indicate sites at which nucleotide sequence changes occurred (6). | size=600px }} |
The first recombination event consists in circularizing the linear PCR product (target-gene flanked by one loxP site upstream and one lox66 site downstream, both in the same orientation) in order to form a plasmid. This step results in a circular fragment comprising the target gene and one lox61 recognition site newly created (Fig. 3), it also produce an 8bp loxP fragment that is released. | The first recombination event consists in circularizing the linear PCR product (target-gene flanked by one loxP site upstream and one lox66 site downstream, both in the same orientation) in order to form a plasmid. This step results in a circular fragment comprising the target gene and one lox61 recognition site newly created (Fig. 3), it also produce an 8bp loxP fragment that is released. | ||
| - | The second recombination event occurs between the circularized PCR product and the Plug&Play plasmid (containing the Cre recombinase gene, a T7 promoter, a lox71 site, a stop site and a ampicillin resistance gene), as an integrative recombination. It results in a bigger plasmid with the target gene located in a context ready to be expressed, flanked by one loxP site upstream and one double mutated site downstream. which reduces the chance of the fragment to be subsequently excised (Fig 3). | + | The second recombination event occurs between the circularized PCR product and the Plug&Play plasmid (containing the Cre recombinase gene, a T7 promoter, a lox71 site, a stop site and a ampicillin resistance gene), as an integrative recombination. It results in a bigger plasmid with the target gene located in a context ready to be expressed, flanked by one loxP site upstream and one double mutated site downstream. which reduces the chance of the fragment to be subsequently excised (Fig. 3). |
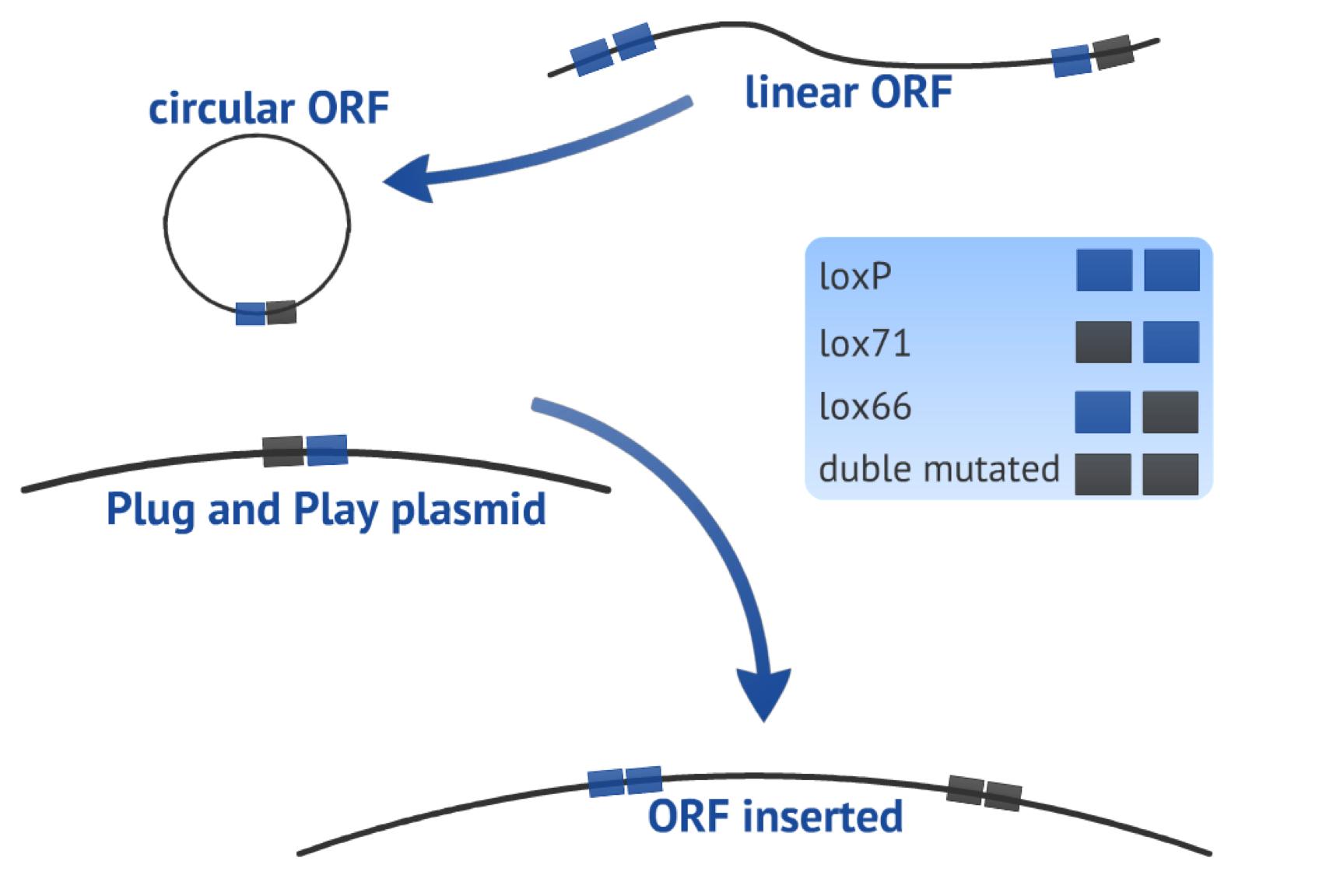
| - | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=fig3plugplay.JPG | caption= Fig.3 ORF circularization and insertion in Plug&Play plasmid | size=600px }} | + | {{:Team:USP-UNESP-Brazil/Templates/RImage | image=fig3plugplay.JPG | caption= Fig.3. ORF circularization and insertion in Plug&Play plasmid | size=600px }} |
This project aims to accomplish the expression of any gene in a two-step process: a PCR reaction and a bacterial transformation. The linear DNA from the PCR is directly inserted into electrocompetent ''E. coli'' cells (e.g. BL21(DE3)) already harboring the Plug&play plasmid and expressing Cre recombinase for the circularization and the integration of the target gene. Mathematical modeling performed to predict rates of linear DNA degradation, integration and circularization showed the viability of the project even before beginning to work in the bench. | This project aims to accomplish the expression of any gene in a two-step process: a PCR reaction and a bacterial transformation. The linear DNA from the PCR is directly inserted into electrocompetent ''E. coli'' cells (e.g. BL21(DE3)) already harboring the Plug&play plasmid and expressing Cre recombinase for the circularization and the integration of the target gene. Mathematical modeling performed to predict rates of linear DNA degradation, integration and circularization showed the viability of the project even before beginning to work in the bench. | ||
Revision as of 10:27, 25 September 2012
 Introduction
Introduction Project Overview
Project Overview Plasmid Plug&Play
Plasmid Plug&Play Associative Memory
Associative MemoryNetwork
 Extras
ExtrasMolecular Mechanism
The Plug&Play project is based on the Cre-lox recombination mechanism, which occurs between the Cre Recombinase enzyme and loxP sites.
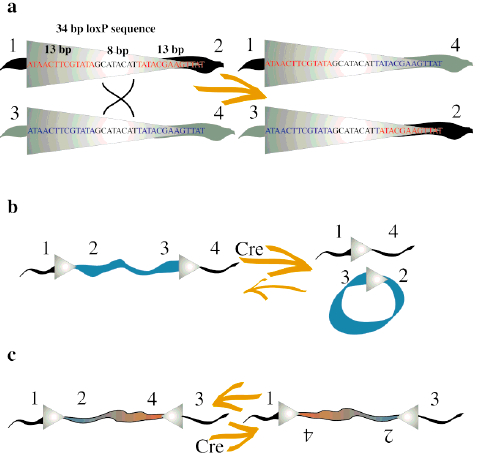
Cre Recombinase (38kDa) is a tyrosine recombinase from the P1 Bacteriophage (1). It plays important roles in this organism’s life cycle, such as circularization of the linear genome when the phage infects the bacteria and resolution of dimeric chromosomes that form after DNA replication (7). The enzyme uses a topoisomeraseI-like mechanism to carry out site-specific recombination events between two DNA recognition sites (loxP sites). Each loxP recognition site has 34bp comprising two 13bp palindromic sequences flanking an 8bp spacer region.
loxP site sequence: ATAACTTCGTATA–GCATACAT–TATACGAAGTTAT
The recombination event is dependent on the location and relative orientation of the loxP sites, which can be in cis (same DNA strand) or trans (different DNA strands) position. The spacer sequence (8bp) found in each loxP site is also important for the outcome of the recombination process. It gives the orientation of one site relative to the other when both are located on the same strand. When two loxP sites are oriented in the same direction the DNA between them will be excised as a circular loop. However, when these two loxP sites are in opposite directions, the DNA between them will be inverted (5) (Fig. 1). The enzyme requires no additional cofactors (such as ATP) or accessory proteins for its function (1).
Concerning to the molecular mechanism of recombination, one recombinase molecule binds to each palindromic half of a loxP site and then the recombinase molecules form a tetramer, thus bringing two loxP sites together (8). The recombination occurs within the spacer area of the loxP sites. The post-recombination loxP sites are formed from the two complementary halves of the pre-recombination sites (Fig. 1). This mechanism was further improved for science research purposes and nowadays lots of tools are based on it. The best-known example is the Nobel Prize winner experiment of genetic knockout on rats using the Cre-lox recombination (4).
Cre-lox recombination between two different DNA fragments can be used for DNA integration. This mechanism has already been extensively explored and it plays a key role in our experiment. Using two DNA fragments, each one with one loxP site, is possible to fuse both into one larger fragment. However, insertion of a circular DNA carrying a loxP into a loxP site on a chromosome (integrative recombination) is quite inefficient because unimolecular reactions are kinetically favored over bimolecular reactions, causing that the inserted DNA will often be excised (6) (Fig. 2a). To tackle this problem we have used loxP mutant sites (2) to help maintaining the inserted DNA in the receptor plasmid (Fig. 2b). In our project we used one loxP site and two loxP mutant sites (lox71 and lox66) (Fig. 2c).
The first recombination event consists in circularizing the linear PCR product (target-gene flanked by one loxP site upstream and one lox66 site downstream, both in the same orientation) in order to form a plasmid. This step results in a circular fragment comprising the target gene and one lox61 recognition site newly created (Fig. 3), it also produce an 8bp loxP fragment that is released.
The second recombination event occurs between the circularized PCR product and the Plug&Play plasmid (containing the Cre recombinase gene, a T7 promoter, a lox71 site, a stop site and a ampicillin resistance gene), as an integrative recombination. It results in a bigger plasmid with the target gene located in a context ready to be expressed, flanked by one loxP site upstream and one double mutated site downstream. which reduces the chance of the fragment to be subsequently excised (Fig. 3).
This project aims to accomplish the expression of any gene in a two-step process: a PCR reaction and a bacterial transformation. The linear DNA from the PCR is directly inserted into electrocompetent E. coli cells (e.g. BL21(DE3)) already harboring the Plug&play plasmid and expressing Cre recombinase for the circularization and the integration of the target gene. Mathematical modeling performed to predict rates of linear DNA degradation, integration and circularization showed the viability of the project even before beginning to work in the bench.
References
1. ABREMSKI & HOESS (1984). "Bacteriophage P1 Site Specific Recombination. Purification and Properties of the Cre Recombinase Protein". Journal of Biological Chemistry 259: 1509–1514.
2. ALBERT H., DALE E.C., LEE E., OW D.W. (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J.7:649–659.
3. HOESS, R.H., ZIESE, M. AND STERNBERG, N. (1982) P1 site-specific recombination: nucleotide sequence of the recombining site. Proc. Nat/Acad. Sci. USA, 79, 3398-3402.
4. MANIS JP. Knock out, knock in, knock down – Genetically manipulated mice and the Nobel Prize. N Engl J Med. 2007; 357:2426
5. NAGY A. 2000. Cre recombinase: the universal reagent for genome tailoring. Genesis 26:99-109.
6. SUZUKI N, INUI M, YUKAWA H (2007) Site-directed integration system using a combination of mutant lox sites for Corynebacterium glutamicum. Appl Microbiol Biotechnol 77: 871-878
7. VAN DUYNE G (2001). "A Structural View of Cre-loxP Site Specific Recombination". Annual Reviews Biophysics Biomolcular Structures 30: 87–104. 8. VOZIYANOV Y, PATHANIA S, JAYARAM M. 1999. A general model for sites pecific recombination by the integrase family recombinases. Nucleic Acids Res 27:930–941
 "
"