Team:USP-UNESP-Brazil/Notebook
From 2012.igem.org
(→First assemblies confirmed) |
(→QR6) |
||
| Line 312: | Line 312: | ||
====QR6==== | ====QR6==== | ||
| + | 08/08/12 – Fernando: 3A Assembly Protocol was used to perform QR5 + 12M + pSB1C3 (QR6) assembly. | ||
| + | 09/08/12 – Fernando: 50uL TOP10 bacteria transformed with 2uL product of the ligation step. | ||
| + | 10/08/12 – Fernando: All colonies were red. | ||
| + | 22/08/12 – Fernando: One more change was made in 3A Assembly Protocol. The enzyme HindIII was used instead of NotI. (This prevented the RFP coding device to ligate again to the pSB1C3 after digestion, so less red colonies were seen). 3A Assembly Protocol was used to perform QR5 + 12M + pSB1C3 (QR6) assembly. | ||
| + | 23/08/12 – Fernando: 50uL TOP10 bacteria transformed with 2uL product of the ligation step. | ||
| + | 26/08/12 – Fernando: Five colonies were inoculated in 5 mL LB + antibiotics and grew overnight. | ||
| + | 27/08/12 – Fernando: DNA was extracted using Quiagen Miniprep Kit. | ||
| + | The QR6 construction was digested with BamHI and PstI, but the 2.363bp and 1.613bp bands could not be distinguished after gel electrophoresis. | ||
| + | 01/09/12 – Fernando: QR6 construction were digested again using a different set of enzymes. 3A Assembly Protocol was used to assemble 12H and 12M using pSB1C3 as plasmid backbone (QR1). | ||
| + | 02/09/12 – Fernando: 50uL TOP10 bacteria transformed with 2uL product of QR1 ligation step. | ||
| + | 03/09/12 – Fernando: All colonies were red | ||
| + | 20/09/12 – Fernando: Since 3A Assembly Protocol was not working to assemble the QR1 construct, each part (12H, 12M and pSB1C3) were digested using the 3A Assembly Protocol digestion procedure and the total volume was loaded in a electrophoresis gel. The idea was to purify the inserts and linearized plasmid backbone pSB1C3 using Quiagen Gel Extraction Kit. Since the bands could barely be seen under U.V, nothing was purified. | ||
====QR1==== | ====QR1==== | ||
Revision as of 03:22, 27 September 2012
 Introduction
Introduction Project Overview
Project Overview Plasmid Plug&Play
Plasmid Plug&Play Associative Memory
Associative MemoryNetwork
 Extras
ExtrasContents
|
Lab Diary
Plasmid Plug&Play
The participation of the people related to the Plug&Play project is show in the Experiments page, it specifies which experiment was performed by each person and the moment (weeks) that it took to be performed.
Associative Memory Network
1- Greg's Lab Experiments
General experiments
17/07/12 – Amanda and Cleandho: - Tetracicline test using TOP10 without plasmid: successful, antibiotic is working
18/07/12 – Aline and Cleandho: - Inoculum of pSB1C3, pSB1K3 and pSB1A2 using RFP as reporter
02/08/12 – Cleandho and Lucas: - Plasmid extraction of pSB1A2, pSB1K3 and pSB1C3 for storage and use. - Make competent cells for storage and use in -80°C
03/08/12 – Cleandho and Lucas: - Competent cells test for viability and competency
22/08 – Cleandho and Lucas: - Make competent cell for use and storage at -80°C - Viability and competency tests of competent TOP 10
23/08/12 – Débora: - Digestion of pSB1K3 with PstI and EcoRI
03/09/12 – Cleandho: - Preparation of LB culture media
05/09/12 – Cleandho and Débora:
- Digestion of pSB1K3, pSB1C3
Assembly experiments
MR2
09/07/12 - Amanda, Cleandho, Débora and Lucas: - Bacterial transformation with MR2 part. MR2 grew colonies
10/07/2012 - Amanda, Cleandho, Débora and Lucas: - Bacterial clones from MR2 were inoculated in LB with tetracycline
11/07/2012 – Amanda, Cleandho, Débora and Lucas: - Plasmid extraction of MR2 and quantification by nanodrop Expected size: MR2 – 25 ng/ul 11J: 82 pb + 12M: 876 pb = 958 pb - Digestion with enzymes and incubate 37 ° C for 1 hour MR2 – (XbaI and PstI)
11/07/2012 – Aline and Cleandho: - Electrophoresis to confirmation: no DNA yield
17/07/12 – Amanda and Cleandho: - Plasmid extraction using BIRNBOIM & DOLY (1979) protocol: failed due non lysis of the cell 18/07/12 – Aline and Cleandho: - Ligation of the parts MR2 using pSB1C3 backbone
19.07.12 – Aline, Cleandho and Débora: - Transformation of MR2
23/07/12 – Cleandho and Débora: - Quantification of pDNA: MR2-2: 119,9 ng/uL - Digestion, using approximately 300ng of DNA MR2: XbaI + PstI
24/07/12 – Cleandho: - Electrophoresis to confirmation of MR2-1 andMR2-2 clones: MR2-2 is positive and MR2-1 was negative.
MR3
09/07/12 - Amanda, Cleandho, Débora and Lucas: - Bacterial transformation with MR3 part. MR3 – grew colonies
10/07/2012 - Amanda, Cleandho, Débora and Lucas: - Bacterial clones from MR3 were inoculated in LB with tetracycline
11/07/2012 – Amanda, Cleandho, Débora and Lucas: - Plasmid extraction of MR3 Expected size: MR3 – 28 ng/ul 2M: 12 pb + 4G: 669 pb = 681 pb - Digestion with enzymes and incubate 37 ° C for 1 hour MR3 – ( EcoRI and SpeI)
11/07/2012 – Aline and Cleandho: - Electrophoresis to confirmation: no DNA yield
18/07/12 – Aline and Cleandho: - Ligation of the part MR3 using pSB1C3 backbone
19.07.12 – Aline, Cleandho and Débora: - Transformation of MR3.
23/07/12 – Cleandho and Débora: - Quantification of pDNA: MR3-1: 110,8 ng/uL - Digestion, using approximately 300ng of DNA MR3: EcoRI + SpeI
24/07/12 – Cleandho: - Electrophoresis to confirmation of MR3-1 clone: no DNA yield
07/08/12 - Cleandho and Lucas: - Digestion of 2M with EcoRI + SpeI and 4G with SbaI + PstI for assembly to MR3
09/08/12 – Amanda - Transformation of MR3 assembly
14/08/12 – Lucas and Amanda - Plasmid extraction from two MR3 clones - Digestion of MR3 clones with (EcoRI + SpeI) - Electrophoresis to confirm MR3 clones: result failed
22/08 – Cleandho and Lucas: - Ligation of assembly MR3 using pSB1K3 (this step was exclusively incubated for 2 hours, all the other ligation steps were incubated overnight) - Transformation of MR3 assembly
04/09/12 – Cleandho: - Plasmid extraction of six MR3 clones - Digestion of MR3 clones with EcoRI and SpeI - Quantification of DNA of MR3 clones: A 178 ng/uL B 129 ng/uL C 119 ng/uL D 98 ng/uL E 159 ng/uL F 110 ng/uL - Digestion of MR3 using HindIII - Electrophoresis of MR3 digested with HindIII. Result: all clones have failed
05/09/12 – Cleandho and Débora: - Digestion of pSB1K3, pSB1C3, 2M, 4G - Ligation of MR3 using 3 proportions of inserts: backbone: 1:1, 3:1 and 1:3
06/09/12 – Cleandho: - Transformation of MR3 assembly in three molar proportions.
11/09/12 – Cleandho and Lucas: - PCR screening of ten colonies clones of assembly MR3
12/09/12 – Cleandho and Lucas: - Electrophoresis of PCR screening clones of MR3 assembly. Result: MR3 shown the expected size of amplicon - Inoculum of clone 4 of MR3 in LBK
13/09/12 Cleandho and Lucas: - Plasmid extraction of clone 4 from MR3 assembly - Digestion of pDNA of clones 4 using EcoRI and SpeI - Electrophoresis for confirmation of clone 4. Result: failed
MR6
09/07/12 - Amanda, Cleandho, Débora and Lucas: - Bacterial transformation MR6 part. MR6 – not grown colonies
17/07/12 – Amanda and Cleandho: - Plasmid extraction using BIRNBOIM & DOLY (1979) protocol: failed due non lysis of the cell
18/07/12 – Aline and Cleandho: - Ligation of the part MR6 using pSB1C3 backbone
19.07.12 – Aline, Cleandho and Débora: - Transformation of MR6
23/07/12 – Cleandho and Débora: - Quantification of pDNA: MR6-1: 45,6ng/uL MR6-5: 46,7ng/uL - Digestion, using approximately 300ng of DNA MR6: EcroRI + SpeI
24/07/12 – Cleandho: - Electrophoresis to confirmation of MR6-1 and MR6-5 clones: all clones have failed
02/08/12 – Cleandho and Lucas: - Ligation/Assembly 3A of MR6 part
03/08/12 – Cleandho and Lucas: - Transformation of MR6
08/08/12 – Débora: - Plasmid extraction from MR6 clones - Digestion of pDNA with EcoRI + PstI from MR6 clones
09/08/12 – Amanda - Electrophoresis to confirm MR6 clones. Result: failed
03/09/12 – Cleandho: - Preparation of LB culture media - Inoculum of six MR6 clones
05/09/12 – Cleandho and Débora: - Digestion of pSB1K3, pSB1C3, 11J and 16P - Ligation of MR6 assembly using 3 proportions of inserts: backbone: 1:1, 3:1 and 1:3
06/09/12 – Cleandho: - Transformation of MR6 assembly in three molar proportions.
12/09/12 – Cleandho and Lucas: - Electrophoresis for reconfirmation of MR6 assembly. Result: MR6 is correct
MR7
14/08/12 – Lucas: - Ligation of assembly MR7 using pSB1K3
24/08/12 – Débora: - Transformation of MR7 assembly
28/08/12 – Débora and Lucas: - Plasmid extraction of MR7 clones - Digestion of MR7 clones with XbaI and PstI
29/08/12 – Débora and Lucas: - Electrophoresis of two clones of MR7 assembly. Result: failed
MR9
14/08/12 – Amanda - Transformation of MR9 assembly
27/08/12 – Lucas: - Plasmid extraction for MR9 clones - Digestion of MR9 clones with EcoRI and SpeI - Electrophoresis of MR9 clones. Result: failed
29/08/12 – Débora and Lucas: - Transformation of MR9 assembly
2- Hotta's and Cleslei's Lab experiments
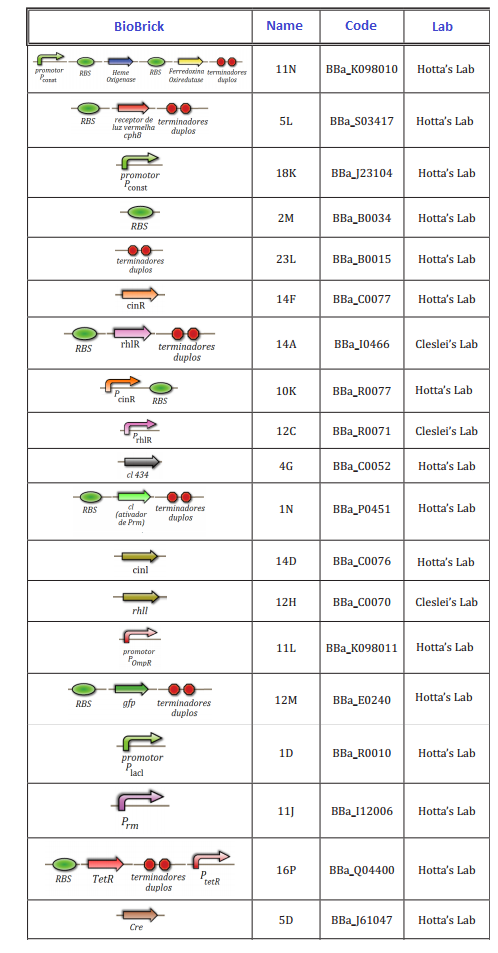
Experiments of Biobricks
At this first moment, we worked on the confirmation of every biobrick that should be used in the project. We transformed and analyzed all the biobricks, using minipreping, nanodroping, digestion and electrophoresis gel. This time was reserved to train everybody in lab techniques, testing protocols and adjusting of the project.
The follow biobricks were confirmed along April and May. On Hotta's Lab participated Lilian, Otto, Daniel, Pedro and Luiza. On Cleslei's Lab, only Fernando
We had issues in the confirmation of small parts (<100 kb), due to the resolution of gel. After this period, we shared the assemblies that should be done in order to start the tests of the system.
Assembly experiments
From 12/06/2012 to 12/08/2012 – Assemblies attempts
At this period, the first attempts in order to construct the lineages that should interact in the system were made. We tried the assemblies QC1, QC6, QR4, MR1 and MR2 (see the full diagram HERE). Nothing worked properly, we had lots of fake positives or assemblies’ transformations that didn’t had any colonies. In order to fix it, we tested every single step of 3A protocols, modifying the proportions of buffer, ligase, vector and parts, assembly time, temperatures and etc. These procedures consumed a big part of our time and resources and the difficulties retarded our advancement, especially on vacations, time when we intent to accelerate the assemblies.
At the end of all tests, we discovered that the enzyme and buffers were, somehow, inadequate. Were made attempts with different brands of buffer and enzymes and a new T4 ligase showed itself as good choice.
First assemblies confirmed
After this messy period trying to fix the assembly's problems, the first good results came. We made with fully success QC1, QC4, QC5, QR4, QR5 and QR1 assemblies through 3A protocol. The results and Agarose Gel photos follows.
QC1
12/06/2012 - Lilian, Ligation
13/06/2012 - Daniel, Transformation
14/06/2012 - Daniel, Inoculate
15/06/2012 - Daniel, Miniprep
16/06/2012 - Otto, Digestion
17/06/2012 - Daniel, Electrophoresis Gel
QC4
06/08/2012 - Pedro, Ligation
07/08/2012 - Pedro, Transformation
10/08/2012 - Daniel, Inoculate
11/06/2012 - Daniel, Miniprep
13/06/2012 - Pedro, Digestion
17/06/2012 - Pedro, Electrophoresis Gel
QC5
14/08/2012 - Daniel, Ligation
QC6
12/06/2012 - Lilian, Ligation
QR4
QR5
QR6
08/08/12 – Fernando: 3A Assembly Protocol was used to perform QR5 + 12M + pSB1C3 (QR6) assembly. 09/08/12 – Fernando: 50uL TOP10 bacteria transformed with 2uL product of the ligation step. 10/08/12 – Fernando: All colonies were red. 22/08/12 – Fernando: One more change was made in 3A Assembly Protocol. The enzyme HindIII was used instead of NotI. (This prevented the RFP coding device to ligate again to the pSB1C3 after digestion, so less red colonies were seen). 3A Assembly Protocol was used to perform QR5 + 12M + pSB1C3 (QR6) assembly. 23/08/12 – Fernando: 50uL TOP10 bacteria transformed with 2uL product of the ligation step. 26/08/12 – Fernando: Five colonies were inoculated in 5 mL LB + antibiotics and grew overnight. 27/08/12 – Fernando: DNA was extracted using Quiagen Miniprep Kit. The QR6 construction was digested with BamHI and PstI, but the 2.363bp and 1.613bp bands could not be distinguished after gel electrophoresis. 01/09/12 – Fernando: QR6 construction were digested again using a different set of enzymes. 3A Assembly Protocol was used to assemble 12H and 12M using pSB1C3 as plasmid backbone (QR1). 02/09/12 – Fernando: 50uL TOP10 bacteria transformed with 2uL product of QR1 ligation step. 03/09/12 – Fernando: All colonies were red 20/09/12 – Fernando: Since 3A Assembly Protocol was not working to assemble the QR1 construct, each part (12H, 12M and pSB1C3) were digested using the 3A Assembly Protocol digestion procedure and the total volume was loaded in a electrophoresis gel. The idea was to purify the inserts and linearized plasmid backbone pSB1C3 using Quiagen Gel Extraction Kit. Since the bands could barely be seen under U.V, nothing was purified.
QR1
18/09/2012 - Daniel & Luiza - Ligation.
At this moment we’re having issues on the assembly of small parts. Other methods of assembling, such as the Standard, are being tested in order to overpass it.
 "
"