Team:USP-UNESP-Brazil/Associative Memory/Experiments
From 2012.igem.org
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:USP-UNESP-Brazil/Templates/Header}} | {{:Team:USP-UNESP-Brazil/Templates/Header}} | ||
| - | + | =Planned Experiments= | |
| - | + | ||
| - | + | ||
| - | + | At first, before doing assembles, we confirmed all parts that were to be used, by verifying the lengths of the inserts in the plasmids using agarose gel and digestion by enzymes that cut in the prefixes and suffixes of BioBricks(EcoRI and PstI). | |
| - | + | Later, we assembled three constructions, each one for an experiment (a test) regarding the genetic device to be used in later constructions. | |
| - | + | ===Tests of "Cin" Quorum Sensing System=== | |
| + | This experiment consists in checking the behavior of Biobricks which make part of the Cin System. We first checked the production of QS substances by observing the enzymatic activity of cinl (enzyme which produce the QS substance), then checked the levels of repression and activation of cinR, and later, the intensity of positive feedback in the system, comparing the levels of activation and inhibition with the previously checked. | ||
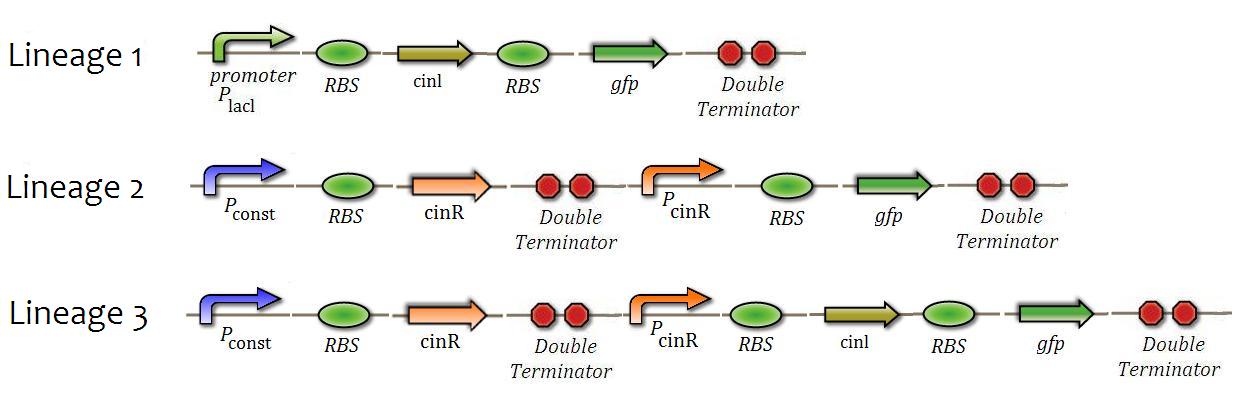
| - | + | Three constructions were made in order to create three different strains of ''E.coli'' (Figure 7). These strains were used in the following tests: | |
| - | + | ||
| - | = | + | [[File:0010.JPG|center|600px|caption=Fig. 7|]] |
| - | + | ===HSL production test=== | |
| - | ===Test of Response to QS | + | On building the ''E.coli'' strain 1, we intended to observe the fluorescence in bacterial growth. This experiment isnot supposed to prove the activity of cinl, but to present signals of its operation, since the GFP would be produced in the same ORF of the cinl gene. <---A graph of fluorescence X IPTG concentration used as input will be made.---> |
| - | + | ||
| - | In the case of any | + | ===Test of Response to QS Substance=== |
| + | ''E.coli'' strain number 2 was built for this test. We intended to centrifuge the medium from the bacteria of strain 1, pouring it on strain 2 and verifying if the members of strain 2 would exhibit fluorescence. Once fluorescence was observed, we would have a strong signal that the QS system was working properly. | ||
| + | |||
| + | In the case of any methodological problem, the GFP from strain 2 can be replaced by RFP. In the case, we would mix strains 1 and 2 members and observe the growth of both simultaneously. If a yellow fluorescence glew (Green + Red) – like on DNA microarrays – we would be sure that strain 2 is producing RFP under influence of strain 1. | ||
===Test for Internal feedback and basal production rate=== | ===Test for Internal feedback and basal production rate=== | ||
| - | In the case of satisfactory results in the previous tests, this one | + | In the case of satisfactory results in the previous tests, this one should probably work as well. The main goal is to estimate if the rate of basal expression is not excessive, in other words, if the positive feedback system is low enough. For this, we can compare the rate of basal fluorescence using the graph made on “HSL production test” as a parameter. |
| - | If the fluorescence | + | If the fluorescence does not appear, there are either the feedback system does not work, or it is too low. In order to verify this issue, the system could be stimulated by insertion of QS substance, measuring the period of glowing and comparing it to the glowing time of bacteria from strain 2 (which does not have a feedback system). |
| - | It is possible to verify how much HSL will be necessary to activate the QS of the system. In order to do this, will be necessary to take samples from the growth medium from | + | It is possible to verify how much HSL will be necessary to activate the QS of the system. In order to do this, it will be necessary to take samples from the growth medium from strain 1 with several different “IPTG inductions”. It is likely to exist a threshold concentration of the QS substance in which the QS starts and the fluorescence of the system keeps glowing without external influence - and our mathematical model suggests this as well. |
| - | ===Tests | + | ===Tests of Rhl system of Quorum Sensing=== |
Just as previously, we intend to check the operation of the QS system, but this time with the rhl system. We will check: production of rhIL enzyme, levels of activation and repression of rhlR substance and ,lastly, intensity positive feedback system, comparing levels of activation and inhibition with tested previously. | Just as previously, we intend to check the operation of the QS system, but this time with the rhl system. We will check: production of rhIL enzyme, levels of activation and repression of rhlR substance and ,lastly, intensity positive feedback system, comparing levels of activation and inhibition with tested previously. | ||
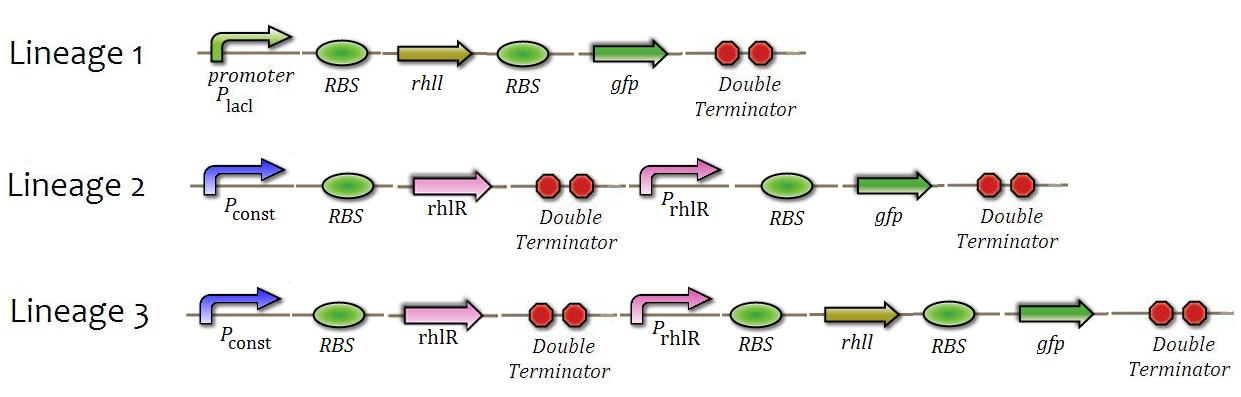
We will do the constructions presented in Figure 8 in addition to tests similar to the system in each Cin. | We will do the constructions presented in Figure 8 in addition to tests similar to the system in each Cin. | ||
| - | [[File:0014.JPG|center|600px|caption|]] | + | [[File:0014.JPG|center|600px|caption=Fig. 8|]] |
| - | + | ||
| + | ===BHL production test=== | ||
| + | The objective of this test is to confirm if the enzyme responsible by the synthesis of QS substance is being produced. Its main evidence is the production of GFP under IPTG input. | ||
| + | <---Using this test, a graph relating the quantity of IPTG and fluorescence will be made in order to be a reference for other tests.---> | ||
| + | |||
| + | ===Reaction to the QS substance test=== | ||
| + | As observed in the test for reaction of Cin system under HSL, a centrifugation of the medium of strain 2 will be poured on bacteria from strain 1. If any issues emerge, the same methodology used on the tests of Cin system should be used. | ||
| - | === | + | ===Test of Internal Feedback and Basal Production Rate Test=== |
| - | + | The levels of fluorescence of strain 3 are to be be compared with the ones of other strains and the concentration of BHL required to activate the QS system is to be be checked. The goals and procedures of this test are equivalent to those in QS Cin system. | |
| - | + | ||
| - | + | ||
| - | Test of Internal Feedback and Basal | + | |
| - | The levels of fluorescence of | + | |
| - | ===Experiments of Multi Regulated Promoter=== | + | ===Experiments of Multi-Regulated Promoter=== |
| - | The main goal of these experiments is checking if the | + | The main goal of these experiments is checking if the BioBricks that constitute the genic regulation system based on the multi-regulated promoter (Prm) are working properly. This promoter is activated by the "cl” factor and inhibited by the “cl434” factor. This system will be used to convert all input signals on activation of the system - indicated by cl production - or on its inhibition of it - indicated by cl434. The main goal is to make sure if the rates of activating or inhibiting of the promoter agree with cl and cl434. |
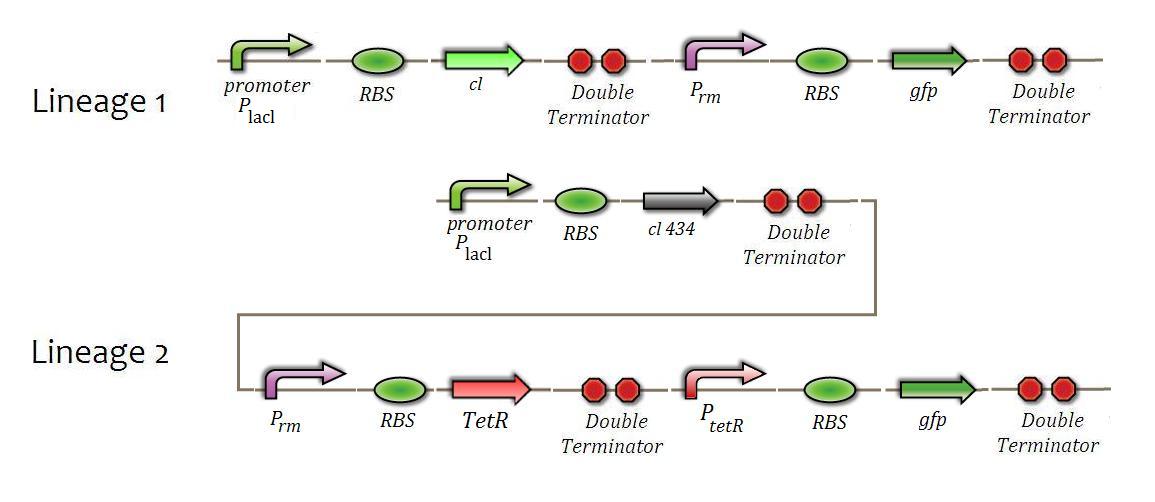
For the Prm promoter system functionality, the follow constructions will be made. | For the Prm promoter system functionality, the follow constructions will be made. | ||
| - | [[File:006.JPG|center|600px|caption|]] | + | [[File:006.JPG|center|600px|caption=Fig. 9|]] |
| - | + | ||
| - | ===Test | + | ===Test of Activation by cl=== |
| - | The main goal of this test is to create the | + | The main goal of this test is to create the “strain 1” described above and induce the production of cl and activation of GFP transcription by use of IPTG. It is expected the appearance of fluorescence, starting in a basal level, in the bacterial growth, |
===Test of Repression by cl434=== | ===Test of Repression by cl434=== | ||
| - | + | ''E.coli'' strain 2, induced by IPTG, produces cl434 and consequently inhibits the production of tetR repressor, which represses the PtetR promoter, the controller of GFP production. In this way, the transcription of GFP by PtetR will be stimulated. Just like in the previous experiment, it is expected to observe the emerging of fluorescence. | |
| - | There is a possibility that the basal levels of Prm transcription | + | There is a possibility that the basal levels of Prm transcription are too high or low to maintain a proper feedback in the QS system. In an ideal situation, the Prm would have transcription rates similar to the QS system promoters (PcinRandPrlhR), emulating the natural feedback of the QS systems- not enough to activate the system. A way to do it would be create mutant Prm promoters with a set of different transcription rates. |
===Assembly diagrams=== | ===Assembly diagrams=== | ||
| - | Previously, | + | Previously, we intended to use a light receptor (Red Light Suit) as input system. The strategy was to put the light responsive promoter in sequence to the Prm promoter, making the genic regulation of the following ORF. In order to simplify the project, this was removed. |
| - | To illustrate all constructions used in all experiments, | + | To illustrate all constructions used in all experiments, a Venn diagram ([https://static.igem.org/mediawiki/2012/2/2a/Assembly_Map.png HERE]) was created, gathering all partial constructions in the main construction. [https://static.igem.org/mediawiki/2012/b/b3/Assemblys_in_english_full.jpg HERE] is the assembly fluxogram. |
| - | We intend to assembly all | + | We intend to assembly all BioBricks using the 3A assembly method, except the smaller parts, like RBS and terminal sequences in which will be used the Standard Assembly Method. Depending on the method of assembly, a different scheme of digestion will be used. The different types of digestion are on the assembly fluxogram. Alternatively to the initial plan using the light switch, IPTG will be used as input to induce the initial production of GFP. |
| - | {{:Team:USP-UNESP-Brazil/Templates/ | + | {{:Team:USP-UNESP-Brazil/Templates/Foot}} |
Latest revision as of 03:49, 27 September 2012
 Introduction
Introduction Project Overview
Project Overview Plasmid Plug&Play
Plasmid Plug&Play Associative Memory
Associative MemoryNetwork
 Extras
ExtrasPlanned Experiments
At first, before doing assembles, we confirmed all parts that were to be used, by verifying the lengths of the inserts in the plasmids using agarose gel and digestion by enzymes that cut in the prefixes and suffixes of BioBricks(EcoRI and PstI). Later, we assembled three constructions, each one for an experiment (a test) regarding the genetic device to be used in later constructions.
Tests of "Cin" Quorum Sensing System
This experiment consists in checking the behavior of Biobricks which make part of the Cin System. We first checked the production of QS substances by observing the enzymatic activity of cinl (enzyme which produce the QS substance), then checked the levels of repression and activation of cinR, and later, the intensity of positive feedback in the system, comparing the levels of activation and inhibition with the previously checked.
Three constructions were made in order to create three different strains of E.coli (Figure 7). These strains were used in the following tests:
HSL production test
On building the E.coli strain 1, we intended to observe the fluorescence in bacterial growth. This experiment isnot supposed to prove the activity of cinl, but to present signals of its operation, since the GFP would be produced in the same ORF of the cinl gene. <---A graph of fluorescence X IPTG concentration used as input will be made.--->
Test of Response to QS Substance
E.coli strain number 2 was built for this test. We intended to centrifuge the medium from the bacteria of strain 1, pouring it on strain 2 and verifying if the members of strain 2 would exhibit fluorescence. Once fluorescence was observed, we would have a strong signal that the QS system was working properly.
In the case of any methodological problem, the GFP from strain 2 can be replaced by RFP. In the case, we would mix strains 1 and 2 members and observe the growth of both simultaneously. If a yellow fluorescence glew (Green + Red) – like on DNA microarrays – we would be sure that strain 2 is producing RFP under influence of strain 1.
Test for Internal feedback and basal production rate
In the case of satisfactory results in the previous tests, this one should probably work as well. The main goal is to estimate if the rate of basal expression is not excessive, in other words, if the positive feedback system is low enough. For this, we can compare the rate of basal fluorescence using the graph made on “HSL production test” as a parameter.
If the fluorescence does not appear, there are either the feedback system does not work, or it is too low. In order to verify this issue, the system could be stimulated by insertion of QS substance, measuring the period of glowing and comparing it to the glowing time of bacteria from strain 2 (which does not have a feedback system).
It is possible to verify how much HSL will be necessary to activate the QS of the system. In order to do this, it will be necessary to take samples from the growth medium from strain 1 with several different “IPTG inductions”. It is likely to exist a threshold concentration of the QS substance in which the QS starts and the fluorescence of the system keeps glowing without external influence - and our mathematical model suggests this as well.
Tests of Rhl system of Quorum Sensing
Just as previously, we intend to check the operation of the QS system, but this time with the rhl system. We will check: production of rhIL enzyme, levels of activation and repression of rhlR substance and ,lastly, intensity positive feedback system, comparing levels of activation and inhibition with tested previously. We will do the constructions presented in Figure 8 in addition to tests similar to the system in each Cin.
BHL production test
The objective of this test is to confirm if the enzyme responsible by the synthesis of QS substance is being produced. Its main evidence is the production of GFP under IPTG input. <---Using this test, a graph relating the quantity of IPTG and fluorescence will be made in order to be a reference for other tests.--->
Reaction to the QS substance test
As observed in the test for reaction of Cin system under HSL, a centrifugation of the medium of strain 2 will be poured on bacteria from strain 1. If any issues emerge, the same methodology used on the tests of Cin system should be used.
Test of Internal Feedback and Basal Production Rate Test
The levels of fluorescence of strain 3 are to be be compared with the ones of other strains and the concentration of BHL required to activate the QS system is to be be checked. The goals and procedures of this test are equivalent to those in QS Cin system.
Experiments of Multi-Regulated Promoter
The main goal of these experiments is checking if the BioBricks that constitute the genic regulation system based on the multi-regulated promoter (Prm) are working properly. This promoter is activated by the "cl” factor and inhibited by the “cl434” factor. This system will be used to convert all input signals on activation of the system - indicated by cl production - or on its inhibition of it - indicated by cl434. The main goal is to make sure if the rates of activating or inhibiting of the promoter agree with cl and cl434.
For the Prm promoter system functionality, the follow constructions will be made.
Test of Activation by cl
The main goal of this test is to create the “strain 1” described above and induce the production of cl and activation of GFP transcription by use of IPTG. It is expected the appearance of fluorescence, starting in a basal level, in the bacterial growth,
Test of Repression by cl434
E.coli strain 2, induced by IPTG, produces cl434 and consequently inhibits the production of tetR repressor, which represses the PtetR promoter, the controller of GFP production. In this way, the transcription of GFP by PtetR will be stimulated. Just like in the previous experiment, it is expected to observe the emerging of fluorescence.
There is a possibility that the basal levels of Prm transcription are too high or low to maintain a proper feedback in the QS system. In an ideal situation, the Prm would have transcription rates similar to the QS system promoters (PcinRandPrlhR), emulating the natural feedback of the QS systems- not enough to activate the system. A way to do it would be create mutant Prm promoters with a set of different transcription rates.
Assembly diagrams
Previously, we intended to use a light receptor (Red Light Suit) as input system. The strategy was to put the light responsive promoter in sequence to the Prm promoter, making the genic regulation of the following ORF. In order to simplify the project, this was removed.
To illustrate all constructions used in all experiments, a Venn diagram (HERE) was created, gathering all partial constructions in the main construction. HERE is the assembly fluxogram.
We intend to assembly all BioBricks using the 3A assembly method, except the smaller parts, like RBS and terminal sequences in which will be used the Standard Assembly Method. Depending on the method of assembly, a different scheme of digestion will be used. The different types of digestion are on the assembly fluxogram. Alternatively to the initial plan using the light switch, IPTG will be used as input to induce the initial production of GFP.
 "
"