|
|
| (17 intermediate revisions not shown) |
| Line 227: |
Line 227: |
| | padding: 0; | | padding: 0; |
| | padding-bottom: 10px; | | padding-bottom: 10px; |
| - | # background:url(http://img.photobucket.com/albums/v26/bluemelon/bg-2-1.jpg); | + | # background:url(https://static.igem.org/mediawiki/2012/c/cc/UCD_Bg_1.jpg); |
| | # background-repeat: no-repeat; | | # background-repeat: no-repeat; |
| | # background-attachment: fixed; | | # background-attachment: fixed; |
| Line 248: |
Line 248: |
| | body { | | body { |
| | background-color: rgba(255,255,255,1); | | background-color: rgba(255,255,255,1); |
| - | background-image: url('http://img.photobucket.com/albums/v26/bluemelon/bg-2-1.jpg'); | + | background-image: url('https://static.igem.org/mediawiki/2012/c/cc/UCD_Bg_1.jpg'); |
| | background-size: 100%; | | background-size: 100%; |
| | background-repeat: no-repeat; | | background-repeat: no-repeat; |
| Line 986: |
Line 986: |
| | <ul> | | <ul> |
| | <li style='color:#014457; cursor:default'><a>teams</a></li> | | <li style='color:#014457; cursor:default'><a>teams</a></li> |
| - | <li class='selected' ><a href="https://2012.igem.org/Team:UC_Davis">Page</a></li> | + | <li class='selected' ><a href="https://2012.igem.org/Team:UC_Davis/Safety">Page</a></li> |
| | | | |
| - | <li class='new'><a href="https://2012.igem.org/wiki/index.php?title=Talk:Team:UC_Davis&action=edit&redlink=1">Discussion</a></li> | + | <li class='new'><a href="https://2012.igem.org/wiki/index.php?title=Talk:Team:UC_Davis/Safety&action=edit&redlink=1">Discussion</a></li> |
| | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UC_Davis/Safety&action=edit">Edit</a></li> | | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UC_Davis/Safety&action=edit">Edit</a></li> |
| | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UC_Davis&action=history">History</a></li> | | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UC_Davis&action=history">History</a></li> |
| - | <li><a href="https://2012.igem.org/Special:MovePage/Team:UC_Davis">Move</a></li> | + | <li><a href="https://2012.igem.org/Special:MovePage/Team:UC_Davis/Safety">Move</a></li> |
| - | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UC_Davis&action=watch">Watch</a></li> | + | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UC_Davis/Safety&action=watch">Watch</a></li> |
| | <li><a href="https://igem.org/Login">Log in</a></li> | | <li><a href="https://igem.org/Login">Log in</a></li> |
| | | | |
| Line 999: |
Line 999: |
| | <div id="newnavi"> | | <div id="newnavi"> |
| | <ul class="newmenu"> | | <ul class="newmenu"> |
| - | <li ><a href="https://2012.igem.org/" title="Back to iGEM">iGEM</a></li>
| + | <li ><a target="new" href="https://2012.igem.org/" title="Back to iGEM">iGEM</a> |
| | + | <ul> |
| | + | <li><a target="new" href="https://2012.igem.org/">Main iGEM</a></li> |
| | + | <li><a href="https://2012.igem.org/Team:UC_Davis/Criteria">Criteria</a></li> |
| | + | <li><a href="https://2012.igem.org/Team:UC_Davis/Human_Practices">Human Practices</a></li> |
| | + | </ul> |
| | + | </li> |
| | | | |
| | <li ><a href="https://2012.igem.org/Team:UC_Davis/Attributions" title="Attributions">Attributions</a></li> | | <li ><a href="https://2012.igem.org/Team:UC_Davis/Attributions" title="Attributions">Attributions</a></li> |
| | | | |
| - | <li ><a href="https://2012.igem.org/Team:UC_Davis/Data" title="Data">Data</a></li> | + | <li ><a title="https://2012.igem.org/Team:UC_Davis/Data" title="Data">Data</a> |
| - | <li ><a href="https://2012.igem.org/Notebook" title="Notebook">Notebook</a>
| + | |
| | <ul> | | <ul> |
| - | <li ><a href="https://2012.igem.org/Team:UC_Davis/Notebook/Overview ">Overview</a></li> | + | <li ><a href="https://2012.igem.org/Team:UC_Davis/Data/Cutinase_Activity" title="Data">Cutinase Activity</a></li> |
| | + | <li ><a href="https://2012.igem.org/Team:UC_Davis/Data/Ethylene_Glycol" |
| | + | title="Data">Ethylene Glycol</a></li> |
| | + | <li ><a href="https://2012.igem.org/Team:UC_Davis/Data/Modeling" |
| | + | title="Data">Modeling</a></li> |
| | + | |
| | + | <li ><a href="https://2012.igem.org/Team:UC_Davis/Parts">Parts</a></li> |
| | + | </ul> |
| | + | </li> |
| | + | <li ><a href="https://2012.igem.org/Team:UC_Davis/Notebook" title="Notebook">Notebook</a> |
| | + | <ul> |
| | + | <li ><a href="https://2012.igem.org/Team:UC_Davis/Notebook">Notebook</a></li> |
| | <li ><a href="https://2012.igem.org/Team:UC_Davis/Notebook/Protocols ">Protocols</a></li> | | <li ><a href="https://2012.igem.org/Team:UC_Davis/Notebook/Protocols ">Protocols</a></li> |
| - | <li ><a href="https://2012.igem.org/Team:UC_Davis/Notebook ">Notebook</a></li>
| |
| | <li ><a href="https://2012.igem.org/Team:UC_Davis/Notebook/Gallery">Gallery</a></li> | | <li ><a href="https://2012.igem.org/Team:UC_Davis/Notebook/Gallery">Gallery</a></li> |
| | </ul> | | </ul> |
| Line 1,018: |
Line 1,033: |
| | <ul> | | <ul> |
| | <li ><a href="https://2012.igem.org/Team:UC_Davis/Project">Project Overview</a></li> | | <li ><a href="https://2012.igem.org/Team:UC_Davis/Project">Project Overview</a></li> |
| - | <li ><a href="https://2012.igem.org/Team:UC_Davis/Project/Catalyst">Modular Engineering</a></li> | + | <li ><a href="https://2012.igem.org/Team:UC_Davis/Project/Catalyst">Module Engineering</a></li> |
| - | <li ><a title="https://2012.igem.org/Team:UC_Davis/Project/Strain">Chassis Engineering</a> | + | <li ><a href="https://2012.igem.org/Team:UC_Davis/Project/Protein_Engineering">Protein Engineering</a></li> |
| | + | |
| | + | <li ><a title="https://2012.igem.org/Team:UC_Davis/Project/Strain">Chassis Engineering </a> |
| | <ul> | | <ul> |
| - | <li><a href="https://2012.igem.org/Team:UC_Davis/Project/Strain">Chassis Engineering</a></li> | + | <li><a href="https://2012.igem.org/Team:UC_Davis/Project/Strain">Background</a></li> |
| | <li><a href="https://2012.igem.org/Team:UC_Davis/Project/Directed_Evolution">Directed Evolution</a></li> | | <li><a href="https://2012.igem.org/Team:UC_Davis/Project/Directed_Evolution">Directed Evolution</a></li> |
| | <li><a href="https://2012.igem.org/Team:UC_Davis/Project/Our_Strain">Rational Engineering </a></li> | | <li><a href="https://2012.igem.org/Team:UC_Davis/Project/Our_Strain">Rational Engineering </a></li> |
| - | </ul></li> | + | </ul> |
| - | <li ><a href="https://2012.igem.org/Team:UC_Davis/Project/Protein_Engineering">Protein Engineering</a></li>
| + | </li> |
| - | <li ><a href="https://2012.igem.org/Team:UC_Davis/Parts">Parts</a></li>
| + | |
| | </ul> | | </ul> |
| | </li> | | </li> |
| Line 1,036: |
Line 1,052: |
| | <div id="slides"> | | <div id="slides"> |
| | | | |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/slide-1-2.jpg" width="850" height="280" alt="" class="current" /> | + | <img src="https://static.igem.org/mediawiki/2012/c/cf/UCD_slide1.jpg" width="850" height="280" alt="" class="current" /> |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/slide-2-2.jpg" width="850" height="280" alt="" /> | + | <img src="https://static.igem.org/mediawiki/2012/e/e3/UCD_Slide_2.jpg" width="850" height="280" alt="" /> |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/slide-3-2.jpg" width="850" height="280" alt="" /> | + | <img src="https://static.igem.org/mediawiki/2012/d/d9/UCD_Slide_3.jpg" width="850" height="280" alt="" /> |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/slide-4-2.jpg" width="850" height="280" alt="" /> | + | <img src="https://static.igem.org/mediawiki/2012/a/a4/UCD_Slide_4.jpg" width="850" height="280" alt="" /> |
| | </div> | | </div> |
| | <ul class="progress"> | | <ul class="progress"> |
| Line 1,116: |
Line 1,132: |
| | | | |
| | <center> | | <center> |
| - | <a href="http://www.fishersci.com" target="_blank"><img src="https://static.igem.org/mediawiki/2011/a/a4/UCD_Fisher_Logo.gif" width="200"></a> | + | <a href="http://www.cs.ucdavis.edu/" target="_blank"><img src="https://static.igem.org/mediawiki/2012/6/6b/UCD_Computer_sponsor.jpg" width="200"></a> |
| | </center> | | </center> |
| | | | |
| | <center> | | <center> |
| | <a href="http://www.bme.ucdavis.edu/" target="_blank"><img src="https://static.igem.org/mediawiki/2011/4/40/UCD_BME_logo_minimal_copy.png" width="200 height="70"></a> | | <a href="http://www.bme.ucdavis.edu/" target="_blank"><img src="https://static.igem.org/mediawiki/2011/4/40/UCD_BME_logo_minimal_copy.png" width="200 height="70"></a> |
| | + | </center> |
| | + | |
| | + | <center> |
| | + | <a href="http://www.fishersci.com" target="_blank"><img src="https://static.igem.org/mediawiki/2011/a/a4/UCD_Fisher_Logo.gif" width="200"></a> |
| | </center> | | </center> |
| | | | |
| Line 1,153: |
Line 1,173: |
| | | | |
| | <div id="myleftbox" class="smallbox"> | | <div id="myleftbox" class="smallbox"> |
| | + | <h1>Background</h1> |
| | + | <article> |
| | + | Originally, we wanted to optimize our engineered bacteria to live in landfills. Our goal was to break down PET at the source of the problem itself. As a team, we took a trip to Yolo County Landfill and learned that a diverse group of bacteria already lives amongst the debris. However, we also learned that our engineered bacteria would have next to no chance of survival amid the competitive microbes already present. Since our <i>E. coli</i> could not survive in the landfill, we shifted our focus to thinking about how the bacteria would work on an industrial level, with the controlled atmosphere of bioreactors to help the <i> E. coli</i> survive. If our project were implemented in industry, this would have the added bonus for safety of containment, which would prevent any concerns associated with releasing the bacteria into the environment. |
| | + | <br> |
| | + | |
| | <h1>Safety</h1> | | <h1>Safety</h1> |
| | <article> | | <article> |
| Line 1,158: |
Line 1,183: |
| | <br><br> | | <br><br> |
| | | | |
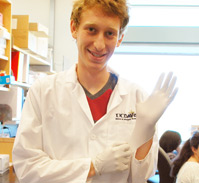
| - | We always made sure to make use of protective gear, such as wearing gloves at all times, masks when using UV lamps, and lab coats when appropriate. All of our waste was autoclaved on site as well, and all tools and glassware thoroughly cleaned after each use. | + | We always made use of protective gear, such as wearing gloves at all times, face masks when using UV lamps, and lab coats when appropriate. All of our waste was autoclaved on site as well, and all tools and glassware thoroughly cleaned after each use. |
| | <br><br> | | <br><br> |
| | | | |
| - | We worked with ethylene glycol during our experiments, which is toxic to the kidney, liver and central nervous system, as well as a skin and eye irritant. Obviously, we wanted to be very safe when using this chemical, so we made sure to consult with our advisers on how best to handle it. When using ethylene glycol, we always worked under a fume hood and wore lab coats. We had a separate waste bottle for all of the byproducts with ethylene glycol, so that the waste could be treated. | + | We worked with ethylene glycol during our experiments, which is toxic to the kidney, liver and central nervous system, as well as a skin and eye irritant. Obviously, we wanted to be very safe when using this chemical, so we made sure to consult with our advisers on how best to handle it. When using ethylene glycol, we always worked under a fume hood and wore lab coats. We had a separate waste bottle for all of the byproducts with ethylene glycol, so that the waste could be treated and properly disposed of. |
| | <br><br> | | <br><br> |
| | | | |
| | <center> | | <center> |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/UCD_sterile3.jpg" align="left"> | + | <img src="https://static.igem.org/mediawiki/2012/3/38/UCD_sterile3.jpg" align="left"> |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/UCD_gloves2-1-1.jpg" align="center"> | + | <img src="https://static.igem.org/mediawiki/2012/2/2e/UCD_gloves2-1.jpg" align="center"> |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/UCD_autoclave3.jpg" align="right"> | + | <img src="https://static.igem.org/mediawiki/2012/3/3d/UCD_autoclave3.jpg" align="right"> |
| | </center> | | </center> |
| | <br> | | <br> |
| Line 1,174: |
Line 1,199: |
| | <br><br> | | <br><br> |
| | | | |
| - | Our BioBricked parts do not raise any safety concerns, as long as the enzymes breaking down ethylene glycol are fully functional.
| |
| - | <br><br>
| |
| | | | |
| - | Our project, when properly functioning, should not have any harmful effect on the environment, researchers, or the public. We keep everything contained within the lab, and all of the plates and wastes are autoclaved to prevent all bacteria we use from spreading to the outside environment. If we were to escalate our project to an industrial level, everything would still be contained within the facility. | + | Our project, when properly functioning, should not have any harmful effect on the environment, researchers, or the public. We keep everything contained within the lab, and all of the plates and wastes are autoclaved to prevent all bacteria we use from spreading to the outside environment. |
| | <br><br> | | <br><br> |
| | | | |
| | If we were to implement our bacteria in a landfill, it could not survive past a couple generations and therefore would not be very toxic. However, if we were to imagine a more robust version of our strain created in the future, we would implement a kill switch, which has already been created using synthetic biology, to make sure we could stop it (2). We would also create a system similar to Imperial College London’s Gene Guard, which would stop to plastic-degrading plasmid from getting transferred to other bacteria (3). This would contain the plasmid within only our bacteria, so it would not get out of hand. | | If we were to implement our bacteria in a landfill, it could not survive past a couple generations and therefore would not be very toxic. However, if we were to imagine a more robust version of our strain created in the future, we would implement a kill switch, which has already been created using synthetic biology, to make sure we could stop it (2). We would also create a system similar to Imperial College London’s Gene Guard, which would stop to plastic-degrading plasmid from getting transferred to other bacteria (3). This would contain the plasmid within only our bacteria, so it would not get out of hand. |
| | <br><br> | | <br><br> |
| - | Our project could potentially be dangerous if our strain received a pathogenic plasmid by lateral gene transfer from other organisms. However, if this were a concern, our strain would be just a susceptible to pathogenicity as any other. If someone with malicious intent were to use our construct, perhaps the most they could do is release it into the water way and have it degrade plastic pipes made of PET, or modify it to degrade other plastics in addition to PET and degrade something useful. Cutinase, the first enzyme in our degradation pathway, functions optimally at 50° C, and the water in our pipes would most likely not be at that temperature. Also, it is unlikely the strain would survive under conditions such as this temperature and being in an aquatic environment. | + | Our project could potentially be dangerous if our strain received a pathogenic plasmid by lateral gene transfer from other organisms. However, if this were a concern, our strain would be just a susceptible to pathogenicity as any other. If someone with malicious intent were to use our construct, perhaps the most they could do is release it into the water way and have it degrade plastic pipes made of PET, or modify it to degrade other plastics in addition to PET and degrade something useful. Cutinase, the first enzyme in our degradation pathway, functions optimally at 50° C, and the water in our pipes would most likely not be at that temperature. Also, it is unlikely the strain would survive under conditions such as this temperature and being in an aquatic environment (4). |
| | <br><br> | | <br><br> |
| | <center> | | <center> |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/UCD_fumehood3.jpg" align="left"> | + | <img src="https://static.igem.org/mediawiki/2012/0/07/UCD_fumehood3.jpg" align="left"> |
| | | | |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/UCD_UV3.jpg" align="center"> | + | <img src="https://static.igem.org/mediawiki/2012/8/8a/UCD_UV3.jpg" align="center"> |
| - | <img src="http://img.photobucket.com/albums/v26/bluemelon/UCD_gloves3.jpg" align="right"> | + | <img src="https://static.igem.org/mediawiki/2012/2/25/UCD_gloves3.jpg" align="right"> |
| | </center> | | </center> |
| | </article> | | </article> |
| Line 1,201: |
Line 1,224: |
| | | | |
| | Our projects involves ethylene glycol, a moderately toxic substance, which is eventually oxidized to oxalic acid – a toxic substance that affects the central nervous system via the liver. Because ethylene glycol must be ingested to pose a problem, researchers take extra precaution to make sure there are no splashes of ethylene glycol in the laboratory and the wastes will be disposed of in the appropriate hazardous waste receptacles. Ethylene glycol can also be a mild irritant if it comes in contact with the skin or if it is inhaled, so researchers wear eye protection as well as gloves and lab coats, and always work with ethylene glycol in the confine of a fume hood. | | Our projects involves ethylene glycol, a moderately toxic substance, which is eventually oxidized to oxalic acid – a toxic substance that affects the central nervous system via the liver. Because ethylene glycol must be ingested to pose a problem, researchers take extra precaution to make sure there are no splashes of ethylene glycol in the laboratory and the wastes will be disposed of in the appropriate hazardous waste receptacles. Ethylene glycol can also be a mild irritant if it comes in contact with the skin or if it is inhaled, so researchers wear eye protection as well as gloves and lab coats, and always work with ethylene glycol in the confine of a fume hood. |
| - | <br>In the environment, ethylene glycol is degraded by hydroxyl radicals and in sewage sludge, it is readily biodegradable. However,ethylene glycol can potentially be toxic within waterways so to ensure environmental safety the team has taken extra precautions to dispose of ethylene glycol safely. | + | <br><br>In the environment, ethylene glycol is degraded by hydroxyl radicals and in sewage sludge, it is readily biodegradable. However, ethylene glycol can potentially be toxic within waterways so to ensure environmental safety the team has taken extra precautions to dispose of ethylene glycol safely. |
| | Additionally, we have considered how to deal with ethylene glycol by designing a circuit in which we produce enzymes that will degrade ethylene glycol into glycoaldehyde and then glycolate. The glycolate has the potential to be turned in to oxaloacetate, a metabolic intermediate. | | Additionally, we have considered how to deal with ethylene glycol by designing a circuit in which we produce enzymes that will degrade ethylene glycol into glycoaldehyde and then glycolate. The glycolate has the potential to be turned in to oxaloacetate, a metabolic intermediate. |
| | | | |
| Line 1,210: |
Line 1,233: |
| | <br> | | <br> |
| | | | |
| - | Our BioBrick parts this year do not raise any serious safety issues. However, One part, the LC-Cutinase gene <a href="http://partsregistry.org/Part:BBa_K936000">BBa_K936000</a>, encodes an enzyme that breaks down PET into ethylene glycol, a moderately toxic substance. This has been noted on its registry page. The safety management procedures have been discussed in the previous question’s answer. The novel part that we are submitting to the Registry is the cutinase gene, in BioBrick format. This is a lipolytic enzyme that does not pose a threat to humans. | + | Our BioBrick parts this year do not raise any serious safety issues. However one part, the LC-Cutinase gene <a href="http://partsregistry.org/Part:BBa_K936000">BBa_K936000</a>, encodes an enzyme that breaks down PET into ethylene glycol, a moderately toxic substance. This has been noted on its registry page. The safety management procedures have been discussed in the previous question’s answer. The novel part that we are submitting to the Registry is the cutinase gene, in BioBrick format. This is a lipolytic enzyme that does not pose a threat to humans. |
| | | | |
| | <br><br> | | <br><br> |
| Line 1,237: |
Line 1,260: |
| | <br>2. Callura, Jared, Dwyer, Daniel, Isaacs, Farren, Cantor, Charles, Collins, James. “Tracking, tuning, and terminating microbial physiology using synthetic riboregulators”. PNAS vol. 107 no. 36, pp. 15898-15903. Web. 13 July 2010. | | <br>2. Callura, Jared, Dwyer, Daniel, Isaacs, Farren, Cantor, Charles, Collins, James. “Tracking, tuning, and terminating microbial physiology using synthetic riboregulators”. PNAS vol. 107 no. 36, pp. 15898-15903. Web. 13 July 2010. |
| | <br>3. “Gene Overview”, <i>Imperial College London iGEM Wiki 2011</i>. | | <br>3. “Gene Overview”, <i>Imperial College London iGEM Wiki 2011</i>. |
| | + | <br>4. S. Sulaiman, S. Yamato, E. Kanaya, J. Kim, Y. Koga, K. Takano, S. Kanaya. "Isolation of a Novel Cutinase Homolog with Polyethylene Terephthalate-Degrading Activity from Leaf-Branch Compost by Using a Metagenomic Approach." Applied and Environment Microbiology, vol. 78 no. 5, pp. 1556-1562, March 2012. |
| | </article> | | </article> |
| | </div> | | </div> |
| Line 1,245: |
Line 1,269: |
| | <!-- site map starts here --> | | <!-- site map starts here --> |
| | <div id="myleftbox" class="smallboxsite"> | | <div id="myleftbox" class="smallboxsite"> |
| - | <ul style="font-size:10px;list-style-image:none;list-style-type:none;float:left;display:inline;color:#000000;" > | + | <ul style="font-size:10px;list-style-image:none;list-style-type:none;float:left;display:inline;color:#000000;" |
| | + | > |
| | + | |
| | + | <li style="float:left;margin:0 10px;"><a |
| | + | href="https://2012.igem.org/Team:UC_Davis"><p>Home</p><ul |
| | + | style="text-indent:-15px; |
| | + | list-style-image:none;list-style-type:none;color:#000000;"><li><a |
| | + | style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis">Welcome</a> </li><li><a |
| | + | style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis">Tweets</a></li><li><a |
| | + | style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis">Sponsors</a> </li><li><a |
| | + | style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Criteria">Criteria</a> </li> |
| | + | </ul> </a> </li> |
| | + | |
| | + | <li style="float:left;margin:0 10px;"><a |
| | + | href="https://2012.igem.org/Team:UC_Davis/Team"><p>Team</p><ul |
| | + | style="text-indent:-15px; |
| | + | list-style-image:none;list-style-type:none;color:#000000;"><li><a |
| | + | style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Team">Who we are</a> |
| | + | </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Team">Students</a></li><li><a |
| | + | style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Team">Advisors</a> </li> |
| | + | </ul> </a> </li> |
| | | | |
| - | <li style="float:left;margin:0 10px solid red"><a href="https://2012.igem.org/Team:UC_Davis"><p>Home</p><ul style="text-indent:-15px; | + | <li style="float:left ;margin:0 10px;"><a |
| - | list-style-image:none;list-style-type:none;color:#000000;"><li>Welcome </li><li>Tweets </li><li>Criteria </li><li>Sponsors </li> </ul> </a> </li> | + | href="https://2012.igem.org/Team:UC_Davis/Project "><p>Project</p></a> |
| | + | <ul style="text-indent:-15px;list-style-image:none;list-style-type:none;color:#000000"><li><a |
| | + | style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Project">Project Overview</a> |
| | + | </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Project/Catalyst ">Module |
| | + | Engineering</a></li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Project/Protein_Engineering |
| | + | ">Protein Engineering</a></li> |
| | + | <li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Project/Strain ">Chassis |
| | + | Engineering</a></li> |
| | + | <li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Project/Directed_Evolution "> |
| | + | - Directed Evolution</a></li> |
| | + | <li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Project/Our_Strain "> - |
| | + | Rational Engineering</a></li> |
| | + | <li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Criteria">Critera</a> |
| | + | </li></ul> </li> |
| | | | |
| - | <li style="float:left ;margin:0 10px;"><a href="https://2012.igem.org/Team:UC_Davis/Team"><p>Team</p> <ul style="text-indent:-15px;list-style-image:none; | + | <li style="float:left ;margin:0 10px"><a |
| - | list-style-type:none;color:#000000 "><li>Who we are </li><li>Students </li><li>Advisors</li></ul></a> </li>
| + | href="https://2012.igem.org/Team:UC_Davis/Safety "> <p>Safety</p></a> |
| | + | <a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Safety "> Safety</a> </li> |
| | | | |
| - | <li style="float:left ;margin:0 10px;"><a href="https://2012.igem.org/Team:UC_Davis/Project "><p>Project</p></a> <ul style="text-indent:-15px;list-style-image:none;list-style-type:none;color:#000000"><li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Project">Project Overview</a> </li><li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Project/Strain ">Modular Engineering</a></li><li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Project/Catalyst ">Chassis Engineering</a></li> | + | <li style="float:left ;margin:0 10px;"><a |
| - | <li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Project/Directed_Evolution "> - Directed Evolution</a></li>
| + | href="https://2012.igem.org/Team:UC_Davis/Notebook "> |
| - | <li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Project/Our_Strain "> - Rational Engineering</a></li> | + | <p>Notebook</p></a> <ul |
| - | <li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Project/Protein_Engineering ">Protein Engineering</a></li>
| + | style="text-indent:-15px;list-style-image:none;list-style-type:none;color:#000000 |
| - | <li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Parts ">Parts</a></li> </ul> </li>
| + | "><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Notebook">Notebook</a> |
| | + | </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Notebook/Protocols |
| | + | ">Protocols</a> </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Notebook/Gallery">Gallery</a> |
| | + | </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Criteria">Critera</a> </li> |
| | + | </ul> </li> |
| | | | |
| - | <li style="float:left ;margin:0 10px"><a href="https://2012.igem.org/Team:UC_Davis/Safety "> <p>Safety</p></a> <a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Safety "> Safety</a> </li> | + | <li style="float:left ;margin:0 10px;"><a |
| | + | title="https://2012.igem.org/Team:UC_Davis/Data "> <p>Data </p></a> <ul |
| | + | style="text-indent:-15px;list-style-image:none;list-style-type:none;color:#000000 |
| | + | "><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Data/Cutinase_Activity "> |
| | + | Cutinase Activity</a> </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Data/Ethylene_Glycol "> |
| | + | Ethylene Glycol</a> </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Data/Modeling "> |
| | + | Modeling</a> </li><li><a style="color:#000000 " |
| | | | |
| - | <li style="float:left ;margin:0 10px;"><a href="https://2012.igem.org/Team:UC_Davis/Notebook "> <p>Notebook</p></a> <ul style="text-indent:-15px;list-style-image:none;list-style-type:none;color:#000000 "><li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Notebook/Overview">Overview</a> </li><li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Notebook/Protocols ">Protocols</a> </li><li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Notebook ">Notebook</a> </li><li><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Notebook/Gallery">Gallery</a> </li> </ul> </li>
| |
| | | | |
| - | <li style="float:left ;margin:0 10px;"><a href="https://2012.igem.org/Team:UC_Davis/Data "> <p>Data </p><ul style="text-indent:-15px;list-style-image:none;
| + | href="https://2012.igem.org/Team:UC_Davis/Parts ">Parts</a></li> </ul> |
| - | list-style-type:none;color:#000000"><li>Data One </li><li>Data Two </li><li>Data Three </li></ul></a> </li>
| + | |
| | | | |
| - | <li style="float:left ;margin:0 10px"><a href="https://2012.igem.org/Team:UC_Davis/Attributions "> <p>Attribution </p></a><a style="color:#000000 " href="https://2012.igem.org/Team:UC_Davis/Attributions "> Attribution</a></li> | + | <li style="float:left ;margin:0 10px"><a |
| | + | href="https://2012.igem.org/Team:UC_Davis/Attributions "> |
| | + | <p>Attribution </p></a><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Attributions "> |
| | + | Attribution</a></li> |
| | | | |
| - | <li style="float:left ;margin:0 10px"><a href="https://2012.igem.org/Main_Page "> <p>iGEM </p></a><a style="color:#000000 " href="https://2012.igem.org/Main_Page " > iGEM</a></li> | + | <li style="float:left ;margin:0 10px"><a |
| - | </ul> | + | href="https://2012.igem.org/Main_Page "> <p>iGEM </p></a><ul |
| | + | style="text-indent:-15px;list-style-image:none;list-style-type:none;color:#000000 |
| | + | "><li><a style="color:#000000 " href="https://2012.igem.org/Main_Page |
| | + | ">Main iGEM</a> </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Criteria "> Criteria</a> |
| | + | </li><li><a style="color:#000000 " |
| | + | href="https://2012.igem.org/Team:UC_Davis/Human_Practices ">Human |
| | + | Practices</a></li> </ul> |
| | </div> | | </div> |
| | <!-- site map ends here --> | | <!-- site map ends here --> |





 "
"











