Team:UC-Merced/Project
From 2012.igem.org
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Notebook | Safety | Background | Attributions |
|---|
Contents |
Overall project
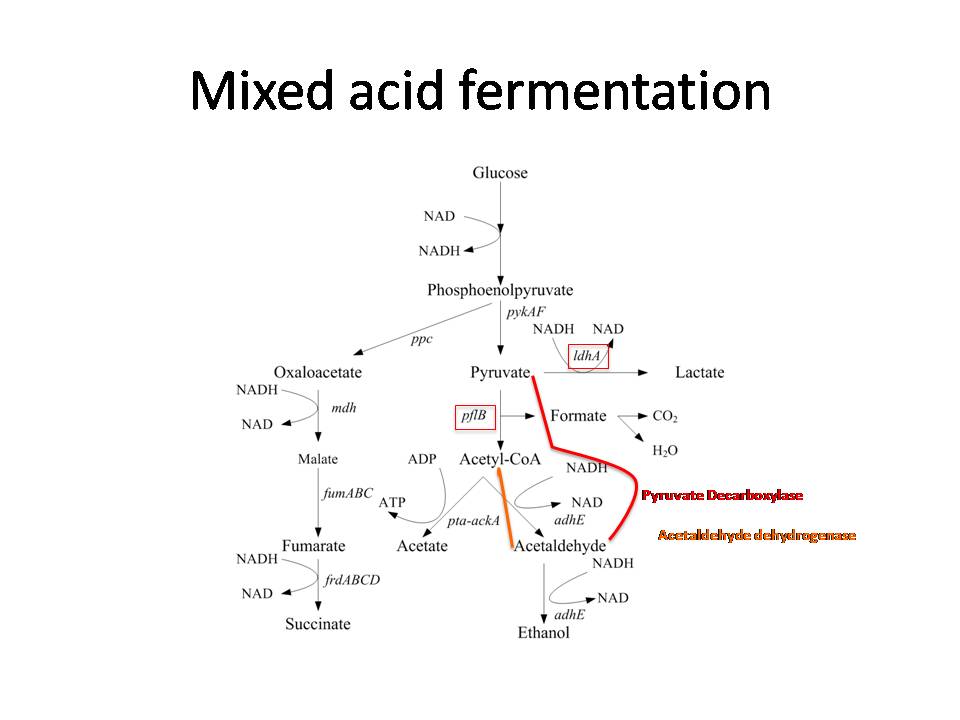
The use of microorganisms as a method to obtain hydrogen gas is well documented but there has yet to be a process which can produce the ideal ratio of glucose to hydrogen gas due to a variety of factors. With each method that has been created, only a select number of pathways are modified in order to produce the desired outcome but never close to the ideal 1:4 mole ratio between glucose and hydrogen gas.
With this as the central focus of our project we are planning to remove pathways involved in dark fermentation and adding an entirely new pathway in the hopes of giving our modified E.coli the ability to generate hydrogen gas as a by product. Furthermore, the new pathway can potentially allow better electron transfer efficiency, which can produce the required 1:4 ratio.
Project Idea
Using NADH ferredoxin oxidoreductase as an electron carrier to shuttle electrons to the hydrogenase to lead to a production in hydrogen gas. Using transduction of the adhE knockout of JW1228-1 to FMJ39 will produce a triple knockout of IdhA, pflB, and adhE. Later, Insertions of mhpF, pyruvate decarboxylase and ferredoxin oxidoreductase will result in a theorhetical production of 4 mol Hydrogen gas per mol of glucose (when the bacteria is placed under fermentation conditions).
Protocols
Magazorb DNA Mini Prep Kit,Promega
Roche Agarose Gel for Electrophoresis
Restriction Enzyme Digest EcoRI
P1 Transduction - Ausubel, F. M. (2001). Current protocols in molecular biology. New York: J. Wiley
Plasmid Transformation [ie. CaCl2 transformation] - Ausubel, F. M. (2001). Current protocols in molecular biology. New York: J. Wiley
Future Work
In our current project, we used NADH ferredoxin oxidoreductase as an electron carrier to shuttle electrons to the hydrogenase and lead to hydrogen production. However, there are other pathways which may yield more hydrogen. In future experiments, we would like to do a qualitative analysis on our system and deduce its efficiency. The bacteria transformed for this project (the FMJ39 strain) may have some pathways which may breakdown hydrogen thus reducing yield. We would like to further study the genome of our microbe and minimize the use of such pathways to maximize our yield.
Our transformed bacteria rely on dark-fermentation to produce hydrogen. However, according to Nath, Kumar, and Das, photo-fermentation is a slightly more advantageous pathway because it is capable of harnessing light energy to drive the reactions in the cell (2005). It also has the advantage of breaking down small organic acids, such as acetyl-CoA in our model, and producing more hydrogen. An efficient system may result from the combination of our model and a photo-fermentation model. Our model is capable of using many compounds and shuttle them to hydrogen production but produces byproducts which cannot be broken down further. The photo-fermentation pathway is capable of using light energy to break down these smaller byproducts and produce more hydrogen.
Our current project took a commonly found fermentation process and modified it to create a funnel for hydrogen production. However, to build a complete and reliable system, we would like to add another system which is capable of producing the substrates needed for the current project. We have identified the breakdown of cellulose, common plant matter, as a potential candidate for this.
Combining a system for cellulose breakdown, dark-fermentation hydrogen production, and photo-fermentation hydrogen production will yield a complete system capable of using raw plant matter to produce hydrogen.
 "
"