Team:Tuebingen/Results
From 2012.igem.org
(Difference between revisions)
Jakobmatthes (Talk | contribs) (→Results) |
|||
| (12 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
__TOC__ | __TOC__ | ||
| - | + | == Parts Status == | |
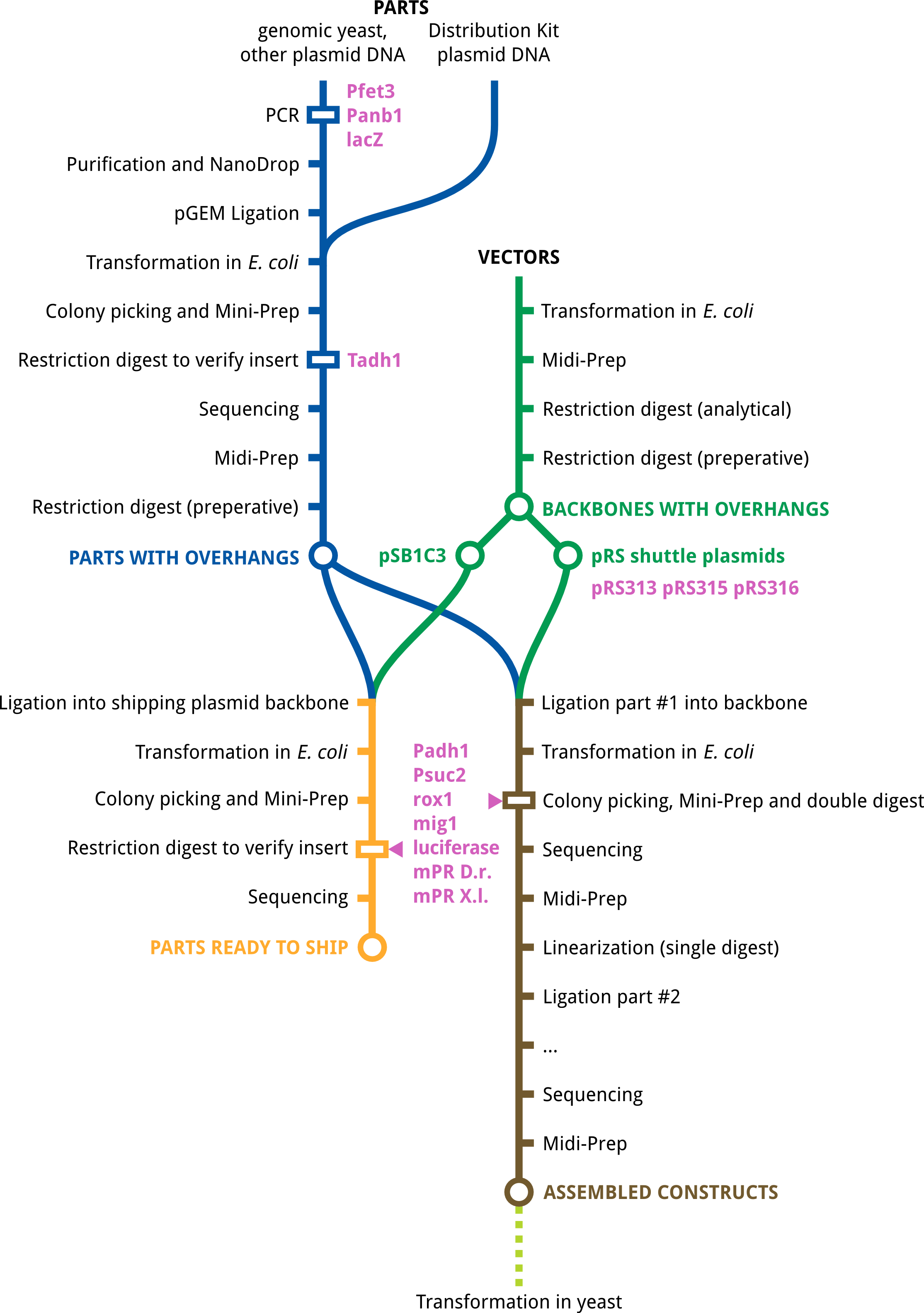
| - | * '''Pfet3, Panb1, lacZ''': PCR was not successful | + | |
| + | * '''Pfet3, Panb1, lacZ''': PCR was not successful after many tries. For Pfet3, new primers were designed but there was no time left to complete this part. | ||
* '''Tadh1''': Tadh1 has been successfully amplified and ligation in pGEM vectors is under way. | * '''Tadh1''': Tadh1 has been successfully amplified and ligation in pGEM vectors is under way. | ||
* '''pRS313, pRS315, pRS316''': The shuttle plasmids are available and digested. | * '''pRS313, pRS315, pRS316''': The shuttle plasmids are available and digested. | ||
| - | * '''Padh1, Psuc2, rox1, mig1, luciferase, mPR ''Danio rerio'', mPR ''Xenopus laevis''''': These parts are all available and digested. The ligation in both pSB1C3 and our pRS shuttle plasmids was | + | * '''Padh1, Psuc2, rox1, mig1, luciferase, mPR ''Danio rerio'', mPR ''Xenopus laevis''''': These parts are all available and digested. The ligation in both pSB1C3 and our pRS shuttle plasmids was not yet successful after many attempts. |
| + | |||
| + | == Identified Problems == | ||
| + | |||
| + | * '''PCR problems:''' Some PCRs yielded only products in very low concentration. Some primers also amplified multiple fragments. New primers partially solved these troubles. | ||
| + | * '''Transformation efficiency:''' The first batch of chemocompetent cells had a very low transformation efficiency. Newly prepared cells showed much better efficiency. | ||
| + | * '''Ligation problems:''' Due to our assembly strategy (only XbaI/SpeI digestion) we had troubles with very low efficiency of insert ligation. Further complication is that the insert can be ligated in the wrong direction. This assembly was necessary because the shuttle plasmids feature only the XbaI/SpeI pair in its multiple cloning site.<br />These problems are the main point why assembly and part submission failed. | ||
| + | * '''Chloramphenicol resistancy and pSB1C3:''' Determining the tolerance of Chloramphenicol of our cells was difficult. Common literature concentrations did not work. In addition to ligation problems, this resulted in no positive pSB1C3 constructs. | ||
| + | |||
| + | == Progress Illustration == | ||
| + | The following illustraion gives a quick overview of the final status of our lab work. | ||
| + | [[File:Tue-labmap-results.png|600px|thumb|center|final status of our parts]] | ||
Latest revision as of 22:54, 26 September 2012
Results
Contents |
Parts Status
- Pfet3, Panb1, lacZ: PCR was not successful after many tries. For Pfet3, new primers were designed but there was no time left to complete this part.
- Tadh1: Tadh1 has been successfully amplified and ligation in pGEM vectors is under way.
- pRS313, pRS315, pRS316: The shuttle plasmids are available and digested.
- Padh1, Psuc2, rox1, mig1, luciferase, mPR Danio rerio, mPR Xenopus laevis: These parts are all available and digested. The ligation in both pSB1C3 and our pRS shuttle plasmids was not yet successful after many attempts.
Identified Problems
- PCR problems: Some PCRs yielded only products in very low concentration. Some primers also amplified multiple fragments. New primers partially solved these troubles.
- Transformation efficiency: The first batch of chemocompetent cells had a very low transformation efficiency. Newly prepared cells showed much better efficiency.
- Ligation problems: Due to our assembly strategy (only XbaI/SpeI digestion) we had troubles with very low efficiency of insert ligation. Further complication is that the insert can be ligated in the wrong direction. This assembly was necessary because the shuttle plasmids feature only the XbaI/SpeI pair in its multiple cloning site.
These problems are the main point why assembly and part submission failed. - Chloramphenicol resistancy and pSB1C3: Determining the tolerance of Chloramphenicol of our cells was difficult. Common literature concentrations did not work. In addition to ligation problems, this resulted in no positive pSB1C3 constructs.
Progress Illustration
The following illustraion gives a quick overview of the final status of our lab work.
 "
"