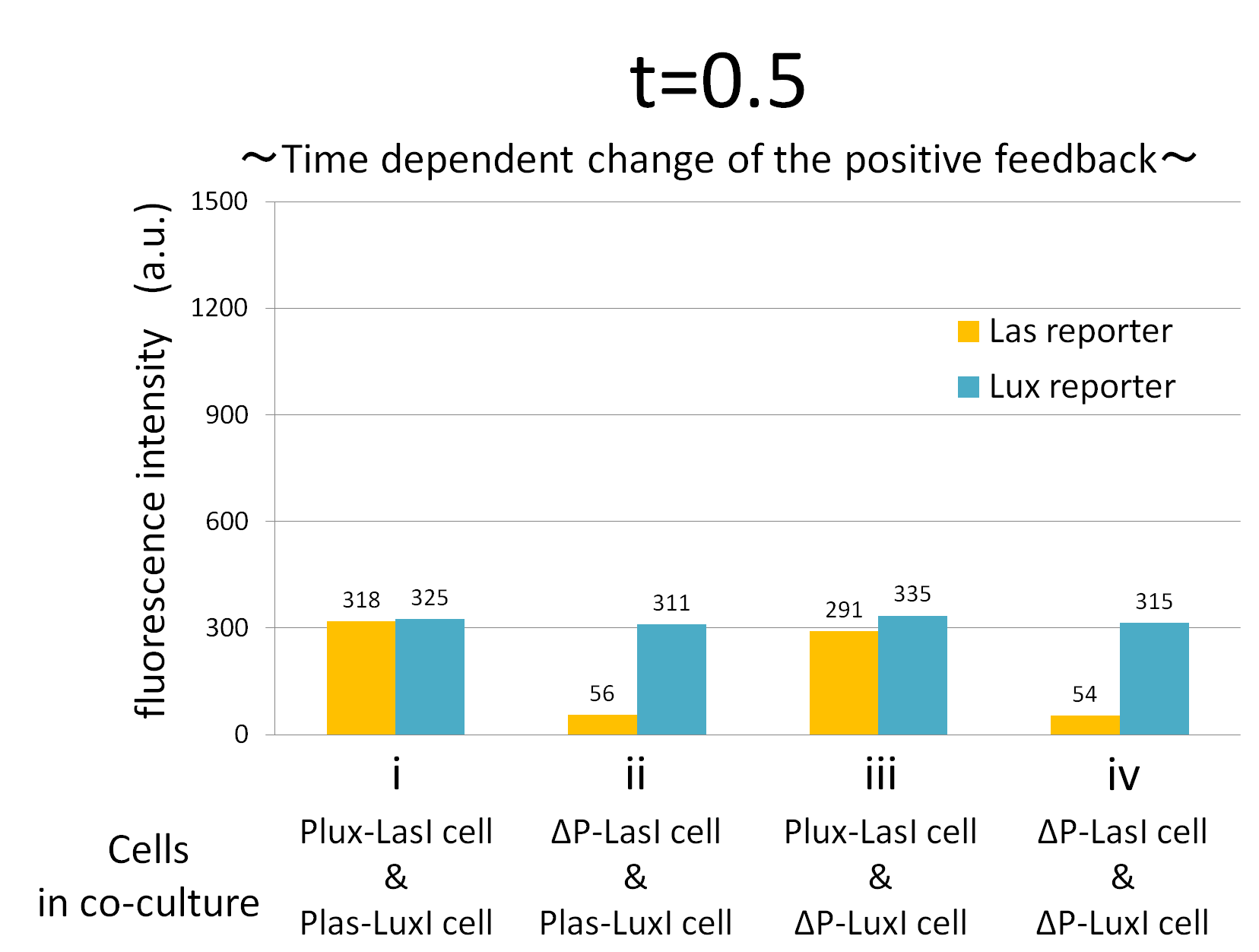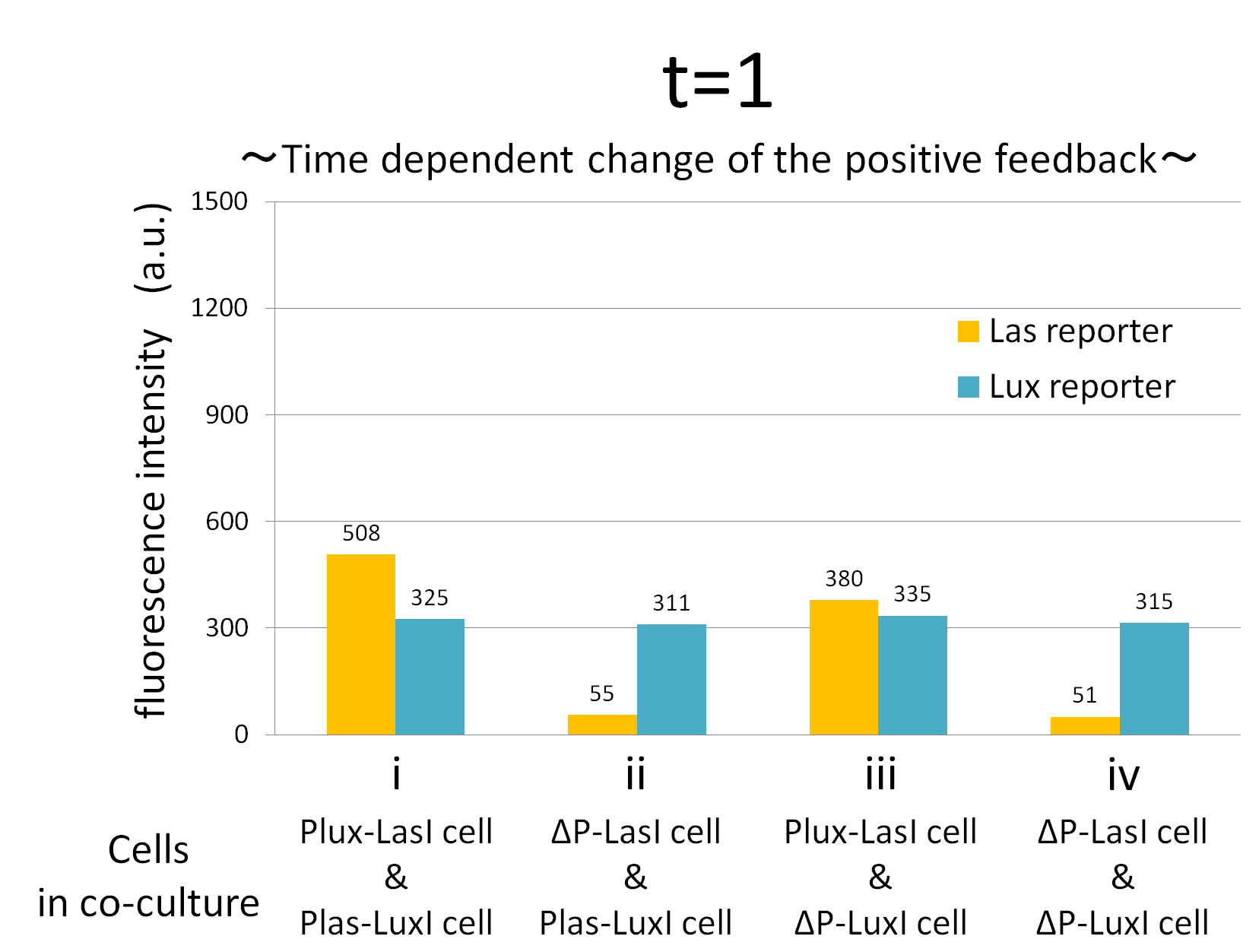Team:Tokyo Tech/positivefeedback2
From 2012.igem.org
Contents |
Time-dependent change assay
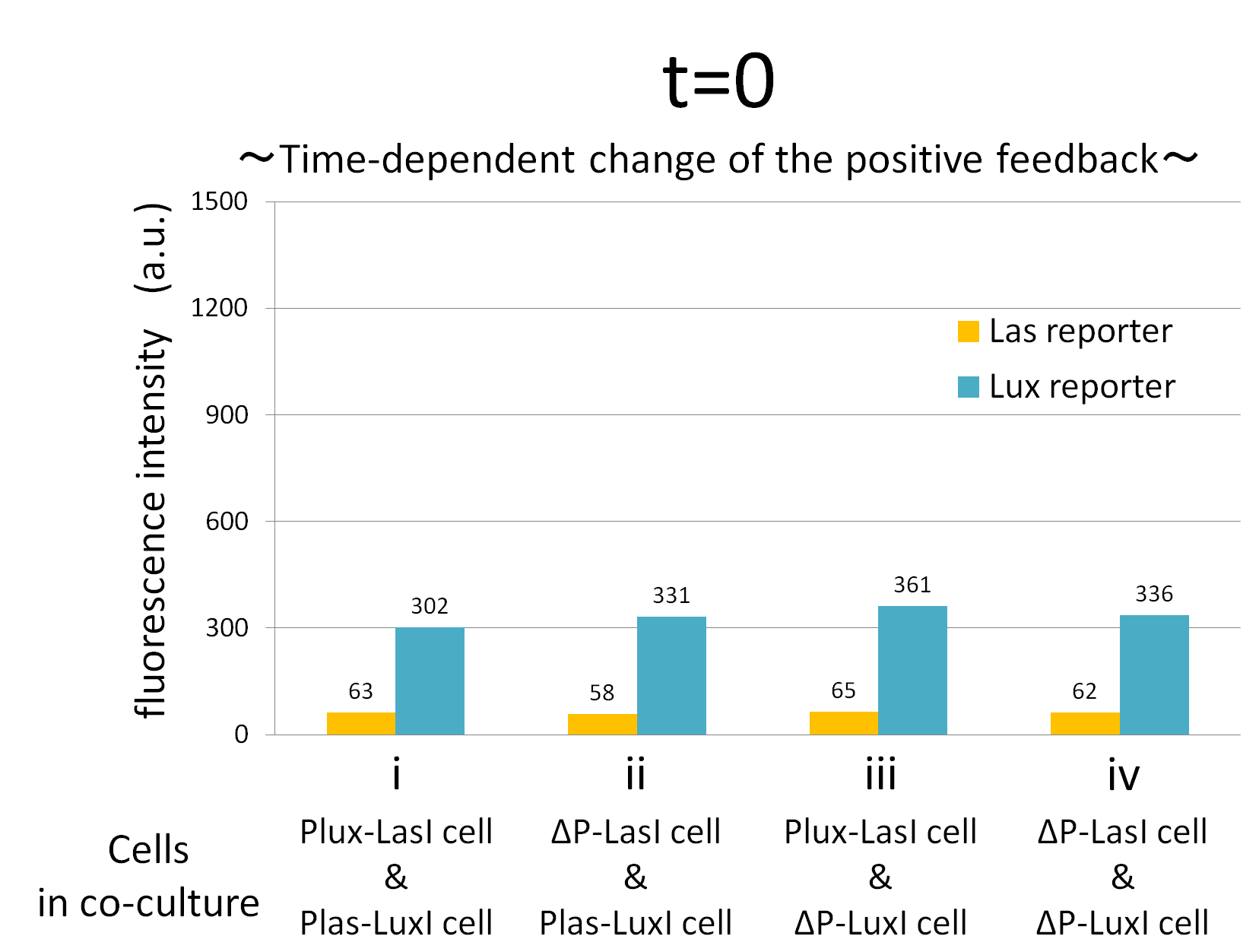
We confirmed the appearance of the positive feedback from the co-culture assay above. To further confirm the positive feedback system, we characterized the temporal behavior of this positive feedback in the cell-cell communication. To observe temporal behavior of the positive feedback, we prepared four conditions of co-culture : Plux-LasI cell and Plas-LuxI cell coexisted in condition i, ΔP-LasI cell and Plas-LuxI cell in condition ii, Plux-LasI cell and ΔP-LuxI in condition iii, and ΔP-LasI and ΔP-LuxI in condition iv. For a trigger of the positive feedback system, we added the initial does of 3OC6HSL (5 nM) to each co-culture.
To estimate the concentration of 3OC6HSL and 3OC12HSL produced by the positive feedback, the signals content in the supernatants of the co-cultures were collected at the time of 0.5, 1, 2, 3, 4hours. The concentration of 3OC6HSL and 3OC12HSL produced by the positive feedback was evaluated by Las reporter strain and Lux reporter strain (Fig2-1-3-1-11). Fig2-1-3-1-11 also shows ideal signal production of the positive feedback in the cell-cell communication.
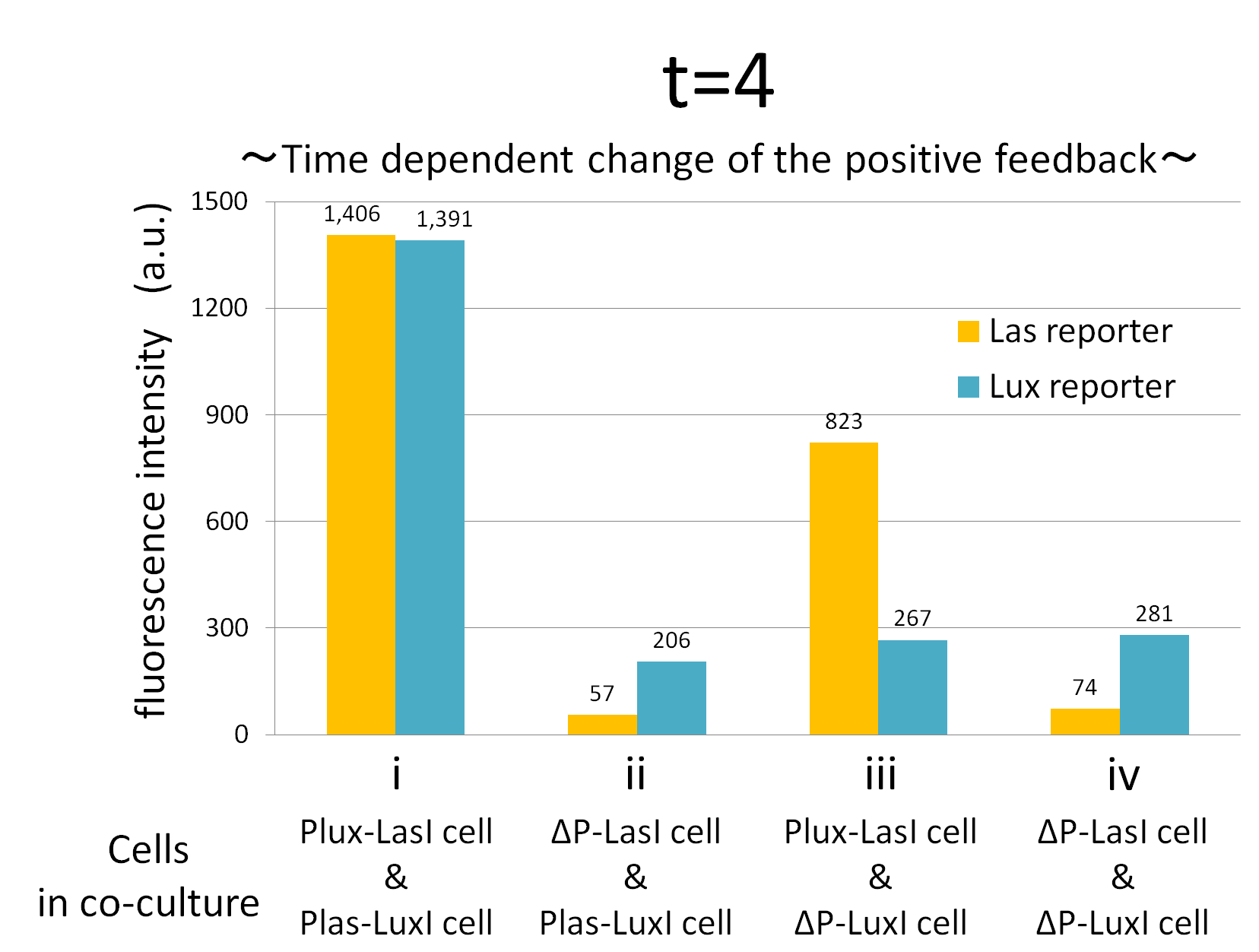
Fig.2-1-3-1-12 shows the characterization of temporal signal increase in the positive feedback in the cell-cell communication. Solid lines represent the fluorescence intensity of Las reporter in the four conditions. Dotted lines represent the fluorescence intensity of Lux reporter in the four conditions.
As compared red solid line with blue dotted line in the condition i (both Plux-LasI cell and Plas-LuxI cell coexist), the fig shows that the fluorescence intensity of Las reporter increases at first (0-0.5h), and then that of Lux reporter starts to increase (1-2h). This result indicates that the 3OC12HSL production in Plux-LasI cell was activated by initially added 3OC6HSL, whereas the 3OC6HSL production in Plas-LuxI cell was not activated till 3OC12HSL production in Plux-LasI cell reached sufficient level. This behavior strongly verifies the appearance of the positive feedback.
Fig2-1-3-1-12 also suggests that the advantage in signal production in the presence of both signal producer (Plux-LasI cell and Plas-LuxI cell). Only when Plux-LasI cell and Plas-LuxI cell were cultured together and allowed to communicate with one another, the fluorescence intensity of Las reporter and Lux reporter increased drastically. On the other hand, when either Plux-LasI cell or Plas-LuxI cell exist (condition ii and iii), drastic rearing in the GFP expression was not observed. Especially in the condition iii, GFP in the las reporter cell is expressed to some extent (about 2-folds lower than in condition i). This behavior shows that initially added 3OC6HSL activated Plux-LasI cell, but the production of 3OC12HSL by Plux-LasI cell in this condition cannot prevail over the production of 3OC12HSL in the positive feedback state. This is because ΔP-LuxI cell cannot produce any 3OC6HSL to activate Plux-LasI cell. Moreover, it goes without saying that, the fluorescence intensity of both Las reporter and Lux reporter remained low level through observation in the absence of both signal producer cell.
In addition, low concentration of 3OC6HSL was detected by the Lux reporter in the condition ii, iii & iv, in which 3OC6HSL is not produced. However, fluorescence intensity in these conditions decreased with time. Therefore, these GFP expressions are derived from initially added 3OC6HSL.
From these overall results, we further confirmed the positive feedback in the cell-cell communication and characterized temporal behavior of the positive feedback. Only when the two types of E.coli were cultured together and allowed to communicate with one another, signal content in the supernatant of the co-culture drastically increase as compared with other conditions.
Materials & Methods
[Go to the project page "Time dependent change assay"]
1,Construction
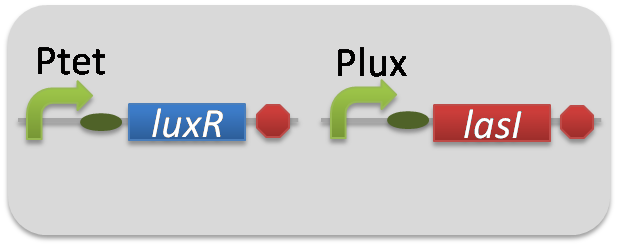
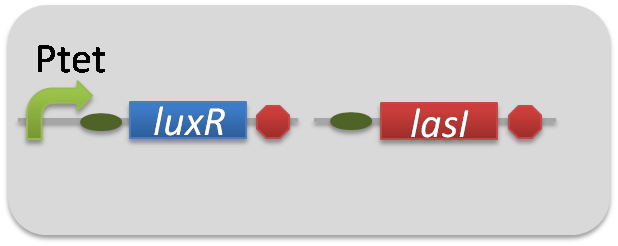
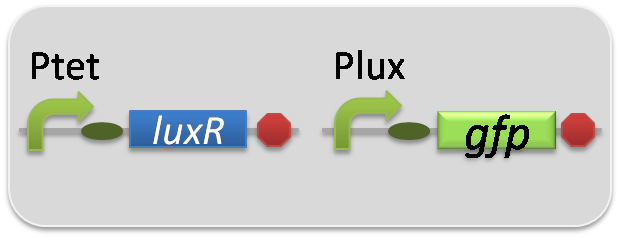
pSB6A1-Ptet-LuxR / pSB3K3-Plux-LasI (JM2.300)…Plux-LasI cell
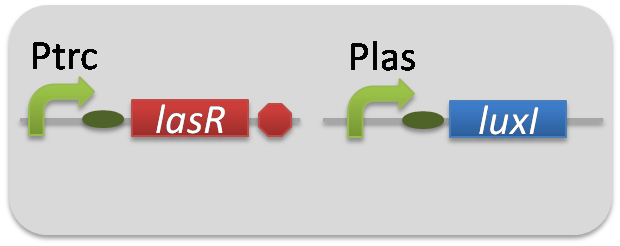
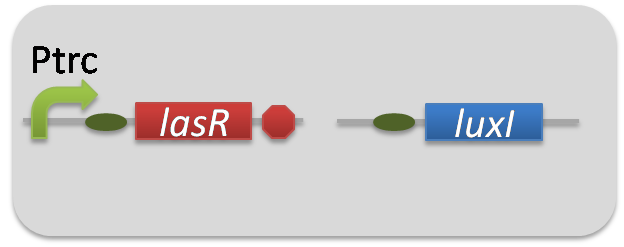
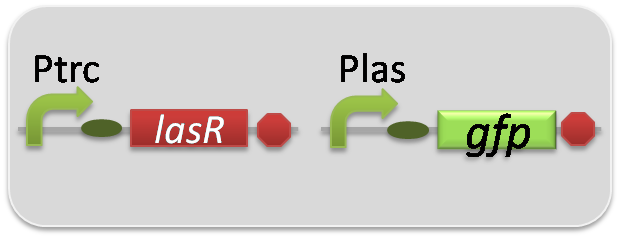
pSB6A1-Ptrc-LasR / pSB3K3-Plas-LuxI (JM2.300)…Plas-LuxI cell
pSB6A1-Ptet-LuxR / pSB3K3-ΔP-LasI (JM2.300)…ΔP-LasI cell
pSB6A1-Ptrc-LasR / pSB3K3-ΔP-LuxI (JM2.300)…ΔP-LuxI cell
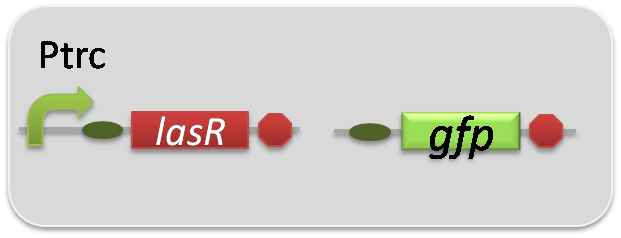
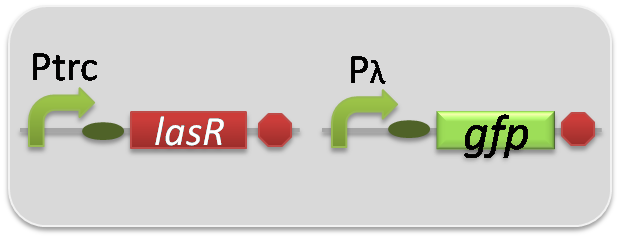
pSB6A1-Ptrc-LasR / pSB3K3-Plas-GFP (JM2.300)…Las reporter cell
pSB6A1-Ptet-LuxR / pSB3K3-Plux-GFP (JM2.300)…Lux reporter cell
pSB6A1-Ptrc-LasR / pSB3K3-ΔP-GFP (JM2.300)…negative control
pSB6A1-Ptrc-LasR / pSB3K3-pλ-GFP (JM2.300)…positive control
2,Strain
JM2.300
Protocol
1. Collect liquid culture
1.1 Prepare overnight culture of inducer cells at 37 °C for 12 hours.
1.2 Take 30 μl of the overnight culture of inducer cells into LB (3 ml) + antibiotics (Amp 50 μg/ml + Kan 30 μg/ml). (→fresh culture)
1.3 Incubate the flesh culture of inducer cells until the observed OD600 reaches around 0.50. Centrifuge the cells at 5000 g, 25 °C, 1 min, suspend it with 1 ml LB + antibiotics (Amp 50 μg/ml + Kan 30 μg/ml).
1.4 According to the Fig, take 30 μl cell suspensions into LB(3 ml) + antibiotics (Amp 50 μg/ml + Kan 30 μg/ml) + 3OC6HSL (5 nM).
1.5 Incubate the inducer cells at 37 °C.
1.6 After incubating 0 ,0.5 ,1 ,2 ,4 hours, centrifuge each inducer cell at 9000 g, 4 °C, 1 min, and filter these cultured cells.
1.7 Samples are moved from round tube to 1.5ml tube and frozen in liquid nitrogen.
1.8 Preserve the samples at -80 °C.
1.9 Dissolve these samples at 37 °C just before process 2.5 ,and dilute the filtrate by LB + antibiotics (Amp + Kan) in 1:30.
2 Reporter assay
2.1 Prepare overnight culture of reporter cells at 37 °C for 12 hours.
2.2 Take 30 μl of the overnight culture of reporter cells into LB (3 ml) + antibiotics (Amp 50 μg/ml + Kan 30 μg/ml). (→fresh culture)
2.3 Incubate the flesh culture of reporter cells until the observed OD600 reaches around 0.50, and gather the supernatant of culture of inducer cells.
2.4 Centrifuge the reporter cells at 5000 g, 25 °C, 1 min, and take it into LB + antibiotics (Amp 50 μg/ml + Kan 30 μg/ml).
2.5 Add 30 μl samples of process 2.4 to filtrate + LB (3 ml) + antibiotics (Amp 50 μg/ml + Kan 30 μg/ml) from process 1.9.
2.6 Incubate the reporter cells for 4 hours at 37 °C.
2.7 Flow cytometer measurements for GFP expression of reporter cells.
 "
"