Team:Tokyo Tech/Projects/PHAs/index.htm
From 2012.igem.org
(→4-1 Confirmation of P(3HB) synthesized on colonies) |
(→4-4 Optimization of the best culture condition to synthesize P(3HB)) |
||
| (80 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
1.</div> | 1.</div> | ||
=Achivement= | =Achivement= | ||
| - | We made a new biobrick part and succeeded in synthesizing Polyhydroxyalkanoates(PHAs). This is the first Biobrick part to synthesize P(3HB), a kind of PHAs. | + | We made a new biobrick part and succeeded in synthesizing Polyhydroxyalkanoates(PHAs). This is the first Biobrick part to synthesize P(3HB), a kind of PHAs. We identified the products as P(3HB) by GC/ MS, and optimised best culture condition to synthesize P(3HB). |
| - | In our project, we also drew rose silhouette to | + | |
| + | In our project, we also drew rose silhouette to reproduce the balcony scene of “Romeo and Juliet” by the synthesis of P(3HB). | ||
<br><br> | <br><br> | ||
<div id="tokyotech" style=" font:bold ;left ; font-size: 30px; color: #0000FF; padding: 2px;"> | <div id="tokyotech" style=" font:bold ;left ; font-size: 30px; color: #0000FF; padding: 2px;"> | ||
| Line 38: | Line 39: | ||
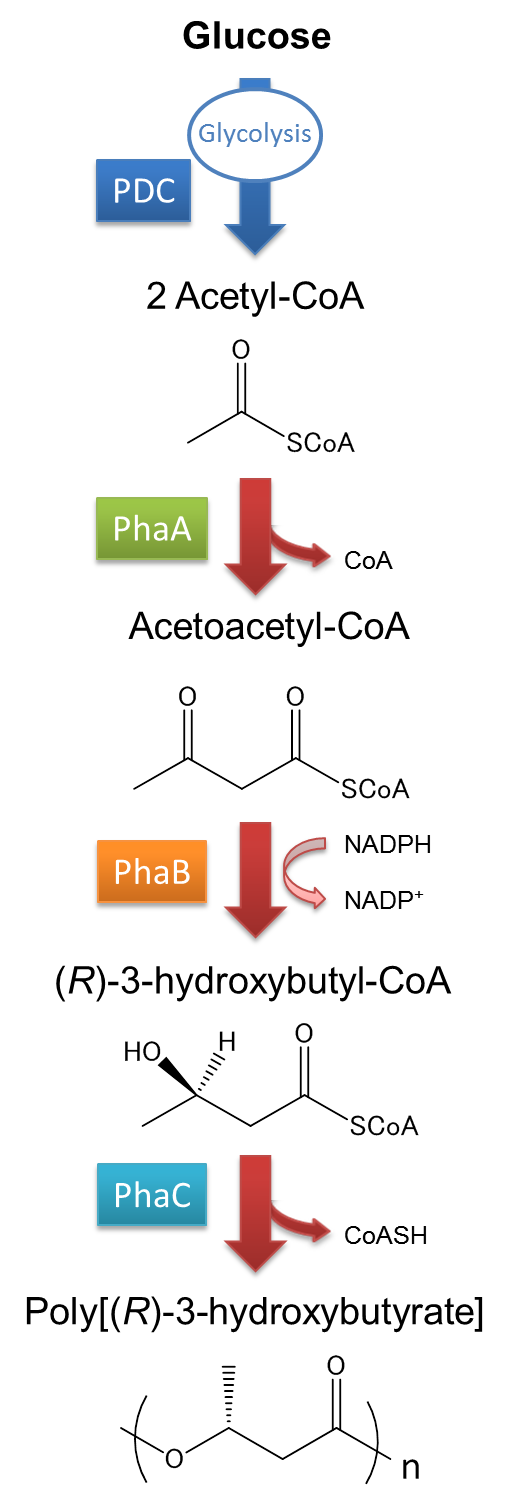
| - | The pathway and regulation of Poly[(R)-3-hydroxybutyrate], P(3HB) synthesis in <I>Ralstonia eutropha</I> H16 is shown in Fig2-2-2-2. Pyruvic acid is metabolized from glucose by glycolysis, and pyruvate dehydrogenase complex (PDC) transforms pyruvic acid into acetyl-CoA. At first, two molecules of acetyl-CoA are ligated to one molecule acetoacetyl-CoA by the action of 3-ketothiolase (coded in PhaA). Acetoacetyl-CoA is transformed into (R)-3-hydroxybutyl-CoA by NADPH dependent acetoacetyl-CoA reductase (coded in PhaB). P(3HB) is then synthesized by the polymerization of (R)-3-hydroxybutyryl-CoA by the action of PHA synthase (PhaC).([[#Reference|[1][2]]] | + | The pathway and regulation of Poly[(R)-3-hydroxybutyrate], P(3HB), synthesis in <I>Ralstonia eutropha</I> H16 is shown in Fig2-2-2-2. Pyruvic acid is metabolized from glucose by glycolysis, and pyruvate dehydrogenase complex (PDC) transforms pyruvic acid into acetyl-CoA. At first, two molecules of acetyl-CoA are ligated to one molecule acetoacetyl-CoA by the action of 3-ketothiolase (coded in PhaA). Acetoacetyl-CoA is transformed into (R)-3-hydroxybutyl-CoA by NADPH dependent acetoacetyl-CoA reductase (coded in PhaB). P(3HB) is then synthesized by the polymerization of (R)-3-hydroxybutyryl-CoA by the action of PHA synthase (PhaC).([[#Reference|[1][2]]] |
) | ) | ||
| Line 59: | Line 60: | ||
To synthesize P(3HB) by <I>E.coli</I>, we transformed <I>E.coli</I> JM109 with the constructed <I>pha C1-A-B1</I> part on pSB1C3 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]). <I>E.coli</I> JM109 is used to synthesize P(3HB), because it tends to have a high density accumulation of P(3HB)([[#Reference|[5]]] | To synthesize P(3HB) by <I>E.coli</I>, we transformed <I>E.coli</I> JM109 with the constructed <I>pha C1-A-B1</I> part on pSB1C3 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]). <I>E.coli</I> JM109 is used to synthesize P(3HB), because it tends to have a high density accumulation of P(3HB)([[#Reference|[5]]] | ||
). As a negative control, we transformed <I>E.coli</I> JM109 with PlasI-gfp on pSB1C3. | ). As a negative control, we transformed <I>E.coli</I> JM109 with PlasI-gfp on pSB1C3. | ||
| + | |||
| + | |||
==4-1 Confirmation of P(3HB) synthesized on colonies== | ==4-1 Confirmation of P(3HB) synthesized on colonies== | ||
| Line 64: | Line 67: | ||
We observed the accumulation of P(3HB) in the <I>E.coli</I> colonies on Nile red positive medium under UV. Nile red has been widely used to stain colonies and distinguish between PHA-accumulating and non-accumulating colonies. Nile red in the agar medium doesn’t affect the growth of the cells, and the accumulation of PHAs in the colonies can be directly monitored([[#Reference|[3][4][5]]] | We observed the accumulation of P(3HB) in the <I>E.coli</I> colonies on Nile red positive medium under UV. Nile red has been widely used to stain colonies and distinguish between PHA-accumulating and non-accumulating colonies. Nile red in the agar medium doesn’t affect the growth of the cells, and the accumulation of PHAs in the colonies can be directly monitored([[#Reference|[3][4][5]]] | ||
). We cultured the transformant on LB agar medium plates with Nile red. After several days, colonies storing P(3HB) were stained orange by Nile red when observed under UV. This result indicates that transformant synthesized and stored P(3HB). | ). We cultured the transformant on LB agar medium plates with Nile red. After several days, colonies storing P(3HB) were stained orange by Nile red when observed under UV. This result indicates that transformant synthesized and stored P(3HB). | ||
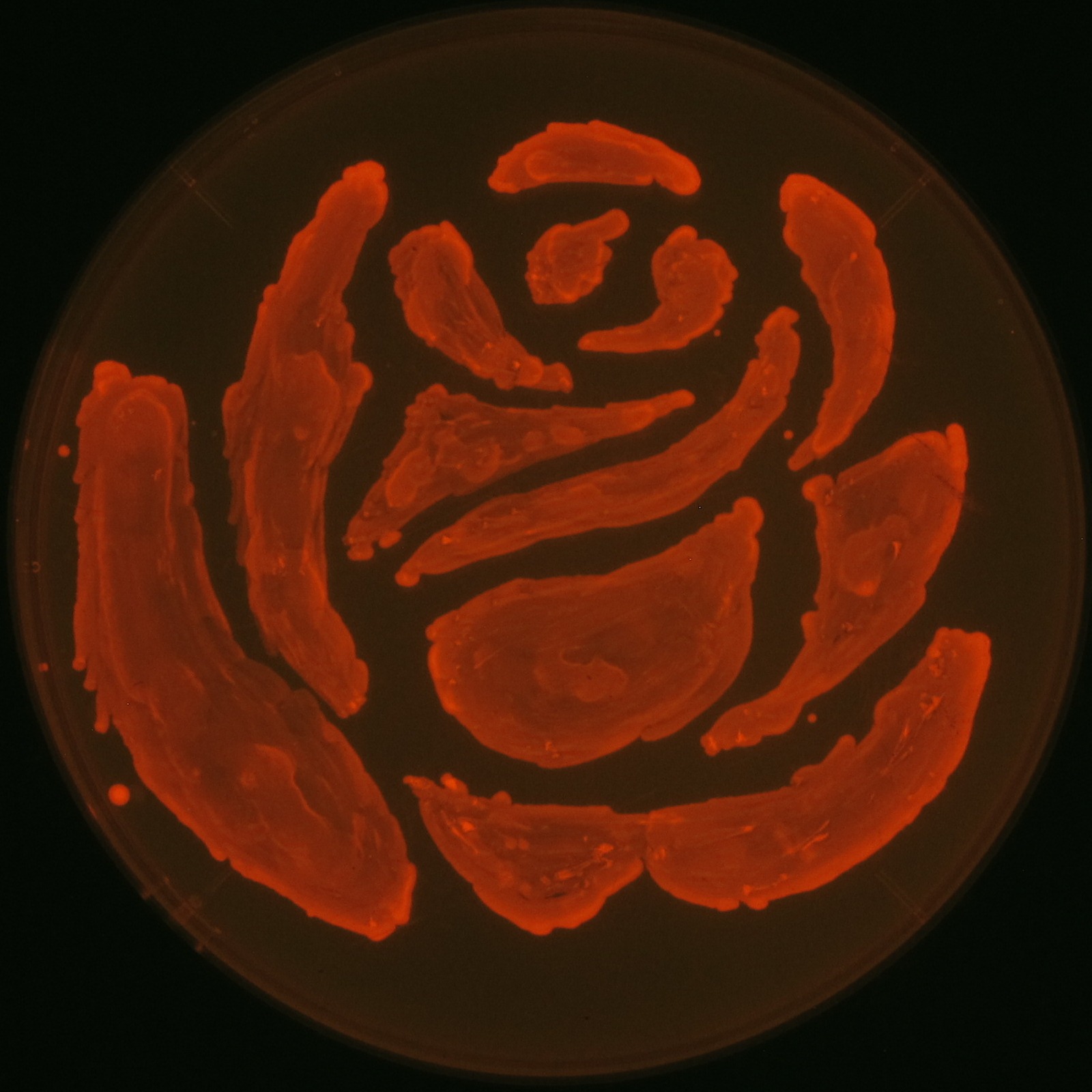
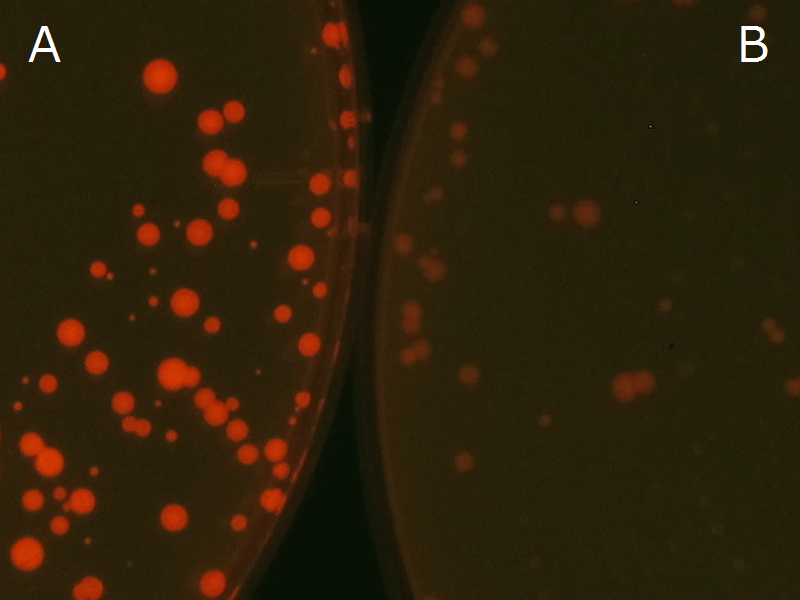
| - | Fig2-2-4-1 is the photographs of <I>E.coli</I> colonies on Nile red positive medium taken under UV. The orange colonies in Fig2-2-4-1A show that the accumulated P(3HB) in cells was stained by Nile red. This result indicates that part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] synthesized P(3HB). Fig2-2-4-1B is the photograph of negative control cells. In this figure we observed that there were no remarkable colored colonies. Fig2-2-4-1-2 shows the difference between cells storing P(3HB) and those not storing P(3HB) on one plate. The cells in blue rectangle area are the cells with P(3HB) synthesis gene and the cells in green rectangle area are the cells with PlasI-gfp gene as a negative control. Using the cells storing P(3HB), we drew a rose silhouette on the LB agar plate containing Nile red (Fig2-2-4-1-3).[[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#A_.P.283HB.29_production_on_colonies_and_preparation_before_confirmation_with_Nile_red_under_UV Protocol]] | + | Fig2-2-4-1-1 is the photographs of <I>E.coli</I> colonies on Nile red positive medium taken under UV. The orange colonies in Fig2-2-4-1-1A show that the accumulated P(3HB) in cells was stained by Nile red. This result indicates that part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] synthesized P(3HB). Fig2-2-4-1-1B is the photograph of negative control cells. In this figure we observed that there were no remarkable colored colonies. Fig2-2-4-1-2 shows the difference between cells storing P(3HB) and those not storing P(3HB) on one plate. The cells in blue rectangle area are the cells with P(3HB) synthesis gene and the cells in green rectangle area are the cells with PlasI-gfp gene as a negative control. Using the cells storing P(3HB), we drew a rose silhouette on the LB agar plate containing Nile red (Fig2-2-4-1-3).[[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#A_.P.283HB.29_production_on_colonies_and_preparation_before_confirmation_with_Nile_red_under_UV Protocol]] |
[[File:tokyotech PHA Nilered1.png|300px|thumb|left|Fig2-2-4-1-1 <br>Fig2-2-4-1-1A: <I>E.coli</I> JM109 colonies with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] gene, PHB accumulation | [[File:tokyotech PHA Nilered1.png|300px|thumb|left|Fig2-2-4-1-1 <br>Fig2-2-4-1-1A: <I>E.coli</I> JM109 colonies with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] gene, PHB accumulation | ||
<br>Fig2-2-4-1-1B: <I>E.coli</I> JM109 colonies with PlasI-gfp gene, no P(3HB) accumulation]] | <br>Fig2-2-4-1-1B: <I>E.coli</I> JM109 colonies with PlasI-gfp gene, no P(3HB) accumulation]] | ||
| Line 76: | Line 79: | ||
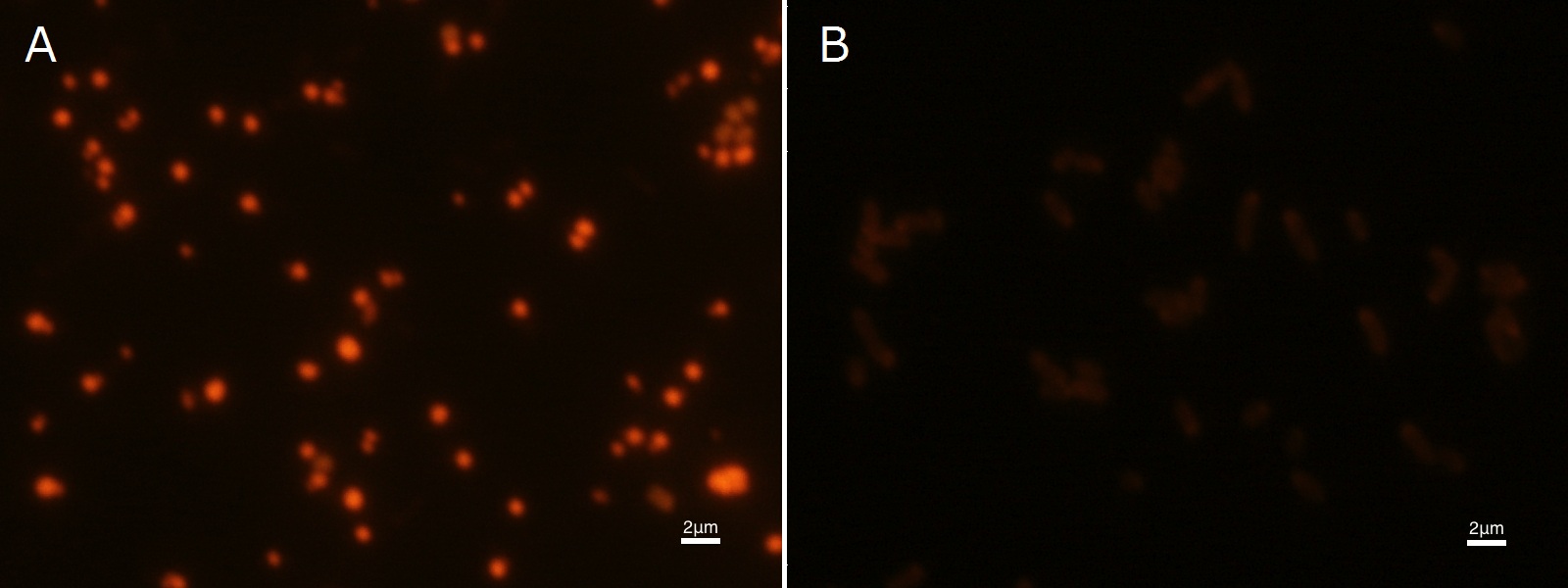
To confirm the accumulation condition of P(3HB) in <I>E.coli</I> with a microscope, we stained the P(3HB) with Nile blue A reagent. Nile blue A is also used to detect the existence of P(3HB) and has no toxicity to the cells([[#Reference|[5]]]). Before the observation, we stained the dried cells with Nile blue A solution. We then took photographs of the sample under fluorescence microscope. | To confirm the accumulation condition of P(3HB) in <I>E.coli</I> with a microscope, we stained the P(3HB) with Nile blue A reagent. Nile blue A is also used to detect the existence of P(3HB) and has no toxicity to the cells([[#Reference|[5]]]). Before the observation, we stained the dried cells with Nile blue A solution. We then took photographs of the sample under fluorescence microscope. | ||
| - | Fig2-2-4-2-1 is the photograph of dried <I>E.coli</I> (with <I>pha C1-A-B1</I> gene) cells dyed with Nile blue A solution taken by fluorescence microscope. The fluorescent areas in Fig2-2-4-2-1A are the accumulated P(3HB) in the cells. This result also indicates that part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] synthesized P(3HB). In the photograph of negative control (Fig2-2-4-2-1B), no remarkable fluorescent area was observed.[[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#B. | + | Fig2-2-4-2-1 is the photograph of dried <I>E.coli</I> (with <I>pha C1-A-B1</I> gene) cells dyed with Nile blue A solution taken by fluorescence microscope. The fluorescent areas in Fig2-2-4-2-1A are the accumulated P(3HB) in the cells. This result also indicates that part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] synthesized P(3HB). In the photograph of negative control (Fig2-2-4-2-1B), no remarkable fluorescent area was observed.[[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#B.P.283HB.29_production_in_cells_and_preparation_before_the_confirmation_with_Nile_blue_A Protocol]] |
[[File:tokyotech PHA Nileblue1.png|800px|thumb|center| | [[File:tokyotech PHA Nileblue1.png|800px|thumb|center| | ||
| Line 82: | Line 85: | ||
Fig2-2-4-2-1B, <I>E.coli</I> JM109 dried cells without P(3HB) accumulation stained by Nile blue A | Fig2-2-4-2-1B, <I>E.coli</I> JM109 dried cells without P(3HB) accumulation stained by Nile blue A | ||
]] | ]] | ||
| + | |||
| + | |||
==4-3 Confirmation of P(3HB) by GC/MS== | ==4-3 Confirmation of P(3HB) by GC/MS== | ||
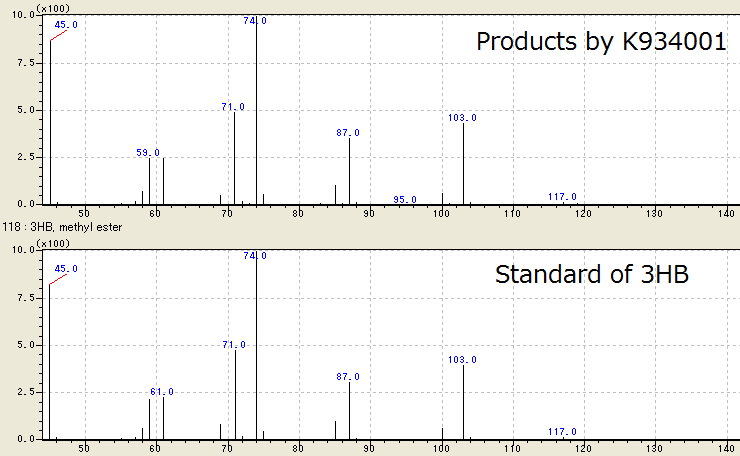
| - | We successfully identified the | + | We successfully identified the products by [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] as 3HB, monomer of P(3HB), by Gas Chromatography/ Mass Spectrometry (GC/ MS). To confirm the products using GC/ MS, the products are methylated because 3HB is difficult to measure. Fig. 2-2-4-3-1 shows the GC/ MS result of the products by [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]. The peaks of sample are same to those of standard control of methylated 3HB. This shows that <I>E.coli</I> synthesized P(3HB) correctly. |
| - | [[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#C. | + | [[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#C._Preparation_for_GC.2FMS Protocol]] |
[[File:tokyotech PHB1.png|800px|thumb|center|Fig2-2-4-3-1, Result of GC/MS]] | [[File:tokyotech PHB1.png|800px|thumb|center|Fig2-2-4-3-1, Result of GC/MS]] | ||
| - | |||
| - | + | ==4-4 Optimization of the best culture condition to synthesize P(3HB)== | |
| - | + | ||
| - | + | ||
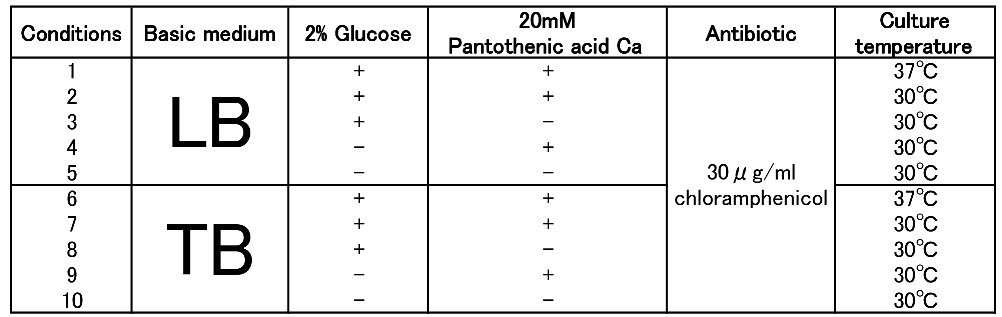
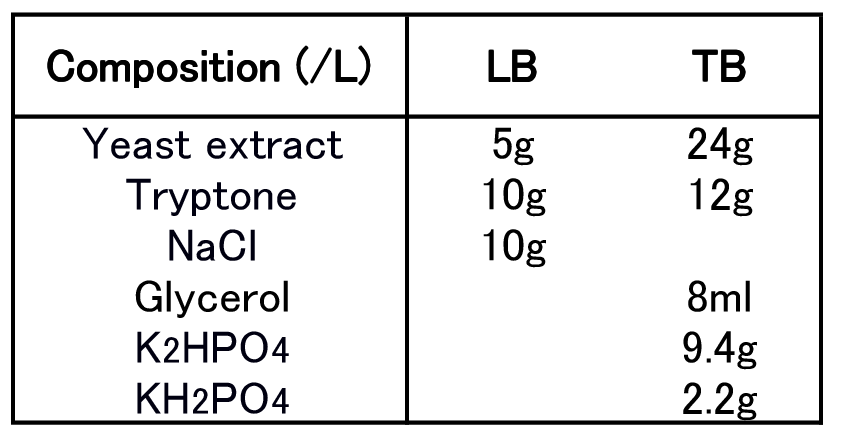
| + | To figure out best culture condition, we tried culturing <I>E.coli</I> JM109 in 10 different conditions for 48h. Each condition is shown in Fig.2-2-4-4-1. Composition of LB and TB medium is shown in Fig. 2-2-4-4-2. | ||
| - | [[File:tokyotech | + | [[File:tokyotech PHB9.png|500px|thumb|left|Fig2-2-4-4-1, different conditions]] |
| + | <br><br> | ||
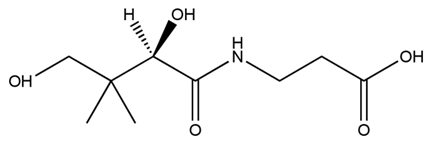
| + | Pantothenic acid (PA), also called vitamin B5 is required to synthesize coenzyme A (CoA). If the glycolytic pathway has become a rate-limiting step, P(3HB) synthesis would be more efficiently by adding PA. | ||
| + | <br><br><br> | ||
| - | + | [[File:tokyotech PHB2.png|380px|thumb|left|Fig2-2-4-4-2, Composition of LB & TB]] | |
| - | + | <br><br> | |
| + | [[File:tokyotech PHB3.png|400px|thumb|right|Fig2-2-4-4-3, Structure of Pantothenic acid | ||
| + | ]] | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br> | ||
| - | |||
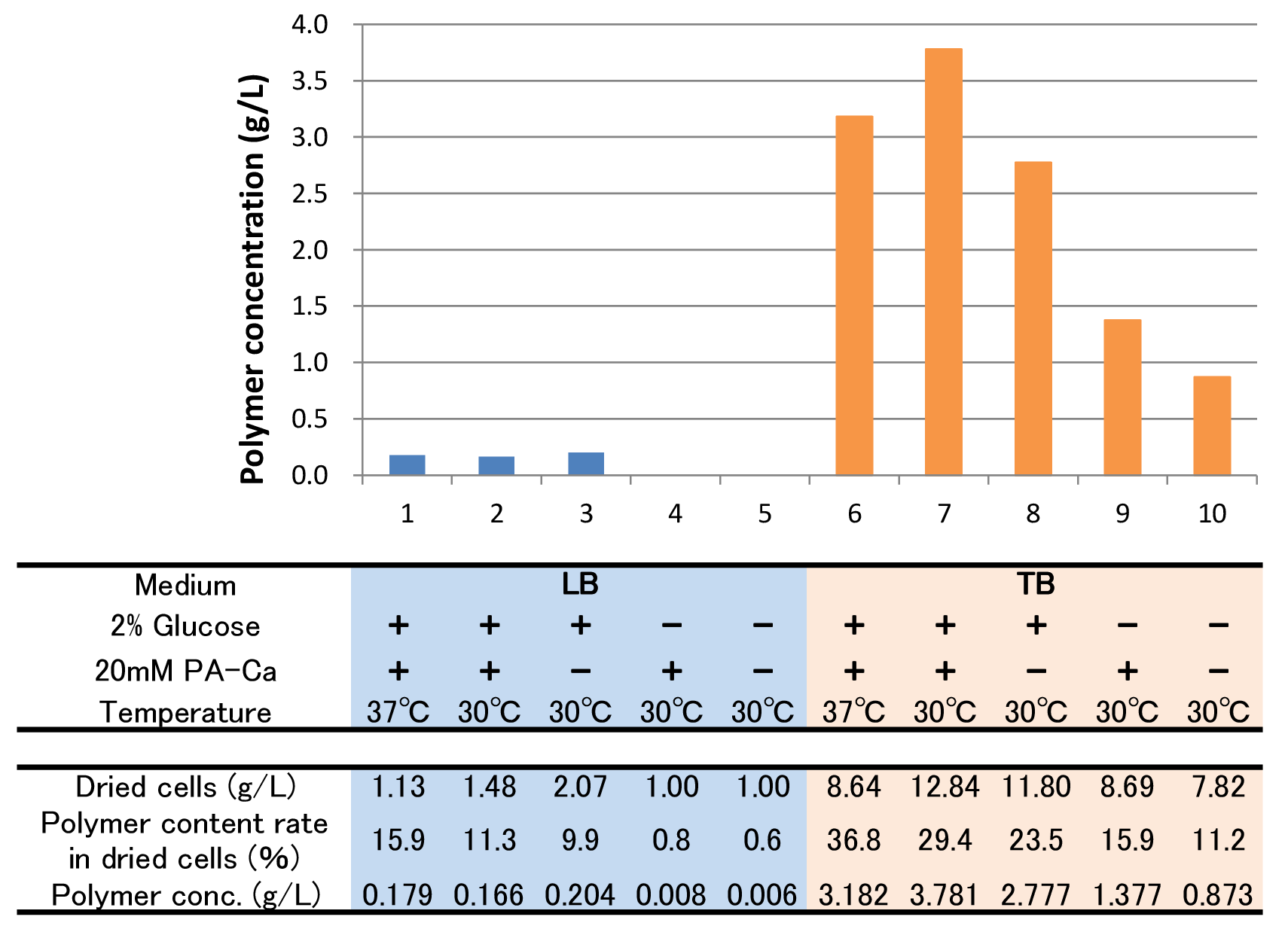
| - | + | The culture result is shown in Fig. 2-2-4-4-4. | |
| - | [[File:tokyotech | + | [[File:tokyotech PHB4.png|800px|thumb|center|Fig2-2-4-5-4, Culture results of ten conditions]] |
| - | ]] | + | |
| - | + | *“Dried cells (g/L)” is the amount of the cells in the medium after culturing. | |
| + | |||
| + | *“Polymer content rate (%)” is the rate of the polymer in the dried cells. | ||
| + | |||
| + | *“Polymer concentration (g/L)” is the amount of the polymer in the medium after culturing. This value is calculated by multiplying “Dried cells” and “Polymer content rate”. | ||
| + | |||
| + | |||
| + | The results showed that TB medium was much better than LB medium to synthesize P(3HB). In both LB and TB, in the 37°C culturing containing glucose and PA-Ca, <I>E.coli</I> synthesized the polymer in maximum content rate. However, the growth of <I>E.coli</I> in 37°C was worse than that in 30°C, therefore final polymer concentration in 37°C and 30°C didn’t make a significant difference. Even if there was no glucose, <I>E.coli</I> synthesized polymer (condition 9 & 10). We think that TB medium had glycerol and a lot of yeast extra, and then <I>E.coli</I> might have used them as carbon sources. In addition, the comparison of condition 4 & 5 indicates PA-Ca was not used as carbon sources. LB medium didn’t contain many carbon sources, so <I>E.coli</I> synthesized little polymer. In this case, adding PA-Ca didn’t have big effect. On the other hand TB medium contains enough carbon sources, so we think that the rate-limiting step was the glycolytic pathway. In this case, polymer production would be increased by adding PA-Ca. (the comparison of condition 7 & 8 and 9&10) | ||
| + | [[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#D._Optimization_of_the_best_culture_condition_to_synthesize_P.283HB.29. Protocol]] | ||
| - | |||
| - | |||
<div id="tokyotech" style=" font:bold ;left ; font-size: 30px; color: #0000FF; padding: 2px;"> | <div id="tokyotech" style=" font:bold ;left ; font-size: 30px; color: #0000FF; padding: 2px;"> | ||
| Line 122: | Line 137: | ||
=Application= | =Application= | ||
[[File:tokyotech PHA perspective.png|200px|thumb|right|Fig2-2-5-1, PHA synthesis gene expression spatially manipulated]] | [[File:tokyotech PHA perspective.png|200px|thumb|right|Fig2-2-5-1, PHA synthesis gene expression spatially manipulated]] | ||
| - | The achievement of our project | + | The achievement of our project “P(3HB) Production” is that we registered available P(3HB) synthetic gene in Biobrick parts. We can control the expression of the P(3HB) synthetic gene spatially by using combination of Biobrick parts. What we want to claim as an example of the spatial manipulation of gene expression is water-repellent. A stronger water-repellent requires hydrophobicity as well as the increase in real surface area that can be achieved as ruggedness of P(3HB) adsorbed on particular surface. If we can control the expression of the P(3HB) synthetic gene spatially by using genetic parts which are registered in Biobrick parts, the application of a super water-repellent sheet will become available. |
| - | We can control the expression of the | + | |
| + | |||
| + | We made P(3HB) sheets. Contact angle is an indicator to represent the strength of the water-repellent. The angle shows the physical properties, especially surface tension.When contact angle of sheets is larger than 90°, from Young equation, the sheets would have stronger water-repellent by increasing real surface area. From literature data, contact angle of P(3HB) sheets is about 100°.[[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB2#Protocol Protocol]] | ||
| + | |||
| + | |||
| + | [[File:tokyotech PHB6.png|600px|thumb|center|Fig2-2-5-2, P(3HB) sheet]] | ||
| + | |||
| + | |||
| + | |||
<br> | <br> | ||
<div id="tokyotech" style=" font:bold ;left ; font-size: 30px; color: #0000FF; padding: 2px;"> | <div id="tokyotech" style=" font:bold ;left ; font-size: 30px; color: #0000FF; padding: 2px;"> | ||
| Line 143: | Line 166: | ||
[6] Vladimir K. Vanag, Cross-diffusion and pattern formation in reaction–diffusion systems, Physical Chemistry Chemical Physics(2009), vol 11.897-912 | [6] Vladimir K. Vanag, Cross-diffusion and pattern formation in reaction–diffusion systems, Physical Chemistry Chemical Physics(2009), vol 11.897-912 | ||
| - | [7] Pohlmann A, et al, Genome sequence of the bioplastic-producing "Knallgas" bacterium Ralstonia eutropha H16, Nat Biotechnol 24:1257-62 | + | [7] Pohlmann A, et al, Genome sequence of the bioplastic-producing "Knallgas" bacterium Ralstonia eutropha H16, Nat Biotechnol(2006), 24:1257-62 |
Latest revision as of 03:56, 27 October 2012
Contents |
Achivement
We made a new biobrick part and succeeded in synthesizing Polyhydroxyalkanoates(PHAs). This is the first Biobrick part to synthesize P(3HB), a kind of PHAs. We identified the products as P(3HB) by GC/ MS, and optimised best culture condition to synthesize P(3HB).
In our project, we also drew rose silhouette to reproduce the balcony scene of “Romeo and Juliet” by the synthesis of P(3HB).
What is PHAs?
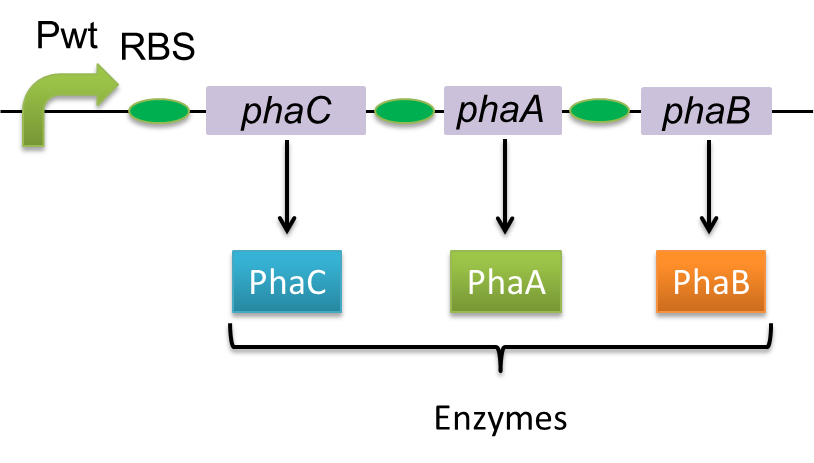
Polyhydroxyalkanoates(PHAs) are biological polyester synthesized by a wide range of bacteria, and can be produced by fermentation from renewable carbon sources such as sugars and vegetable oils. These polyesters are biodegradable thermoplastics and elastomers, which exhibit interesting material properties. PHAs are also a kind of bio plastics, which can be biodegraded a lot faster than fossil-fuel plastics in the environment. Poly-3-hydroxybutyrate, P(3HB) is the most common type of PHAs. P(3HB) is synthesized by the enzymes coded in the gene of PHA synthesis (pha C1-A-B1) from Ralstonia eutropha H16.
Poly-3-hydroxybutyrate, P(3HB) is synthesized by three enzymes.
The A gene encodes for the 393 amino acids protein, 3-ketothiolase (PhaA)
The B gene encodes for the 246 amino acids protein, acetoacetyl-CoA reductase (PhaB)
The C gene encodes for the 589 amino acids protein, PHA Synthase (PhaC)
The pathway and regulation of Poly[(R)-3-hydroxybutyrate], P(3HB), synthesis in Ralstonia eutropha H16 is shown in Fig2-2-2-2. Pyruvic acid is metabolized from glucose by glycolysis, and pyruvate dehydrogenase complex (PDC) transforms pyruvic acid into acetyl-CoA. At first, two molecules of acetyl-CoA are ligated to one molecule acetoacetyl-CoA by the action of 3-ketothiolase (coded in PhaA). Acetoacetyl-CoA is transformed into (R)-3-hydroxybutyl-CoA by NADPH dependent acetoacetyl-CoA reductase (coded in PhaB). P(3HB) is then synthesized by the polymerization of (R)-3-hydroxybutyryl-CoA by the action of PHA synthase (PhaC).([1][2]
)
Construction of phaC1-A-B1 in Biobrick format
In this study, we constructed a part containing phaC1-A-B1 in Biobrick format([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]).[Construction of PHA-C1-A-B1 in Biobrick format]
This is the first Biobrick part which worked as expected though some teams had tried to synthesize PHAs in the past iGEM.[Production trial of PHAs by past teams]
P(3HB) production by E.coli & Confirmation of P(3HB)
To synthesize P(3HB) by E.coli, we transformed E.coli JM109 with the constructed pha C1-A-B1 part on pSB1C3 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]). E.coli JM109 is used to synthesize P(3HB), because it tends to have a high density accumulation of P(3HB)([5] ). As a negative control, we transformed E.coli JM109 with PlasI-gfp on pSB1C3.
4-1 Confirmation of P(3HB) synthesized on colonies
We observed the accumulation of P(3HB) in the E.coli colonies on Nile red positive medium under UV. Nile red has been widely used to stain colonies and distinguish between PHA-accumulating and non-accumulating colonies. Nile red in the agar medium doesn’t affect the growth of the cells, and the accumulation of PHAs in the colonies can be directly monitored([3][4][5] ). We cultured the transformant on LB agar medium plates with Nile red. After several days, colonies storing P(3HB) were stained orange by Nile red when observed under UV. This result indicates that transformant synthesized and stored P(3HB). Fig2-2-4-1-1 is the photographs of E.coli colonies on Nile red positive medium taken under UV. The orange colonies in Fig2-2-4-1-1A show that the accumulated P(3HB) in cells was stained by Nile red. This result indicates that part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] synthesized P(3HB). Fig2-2-4-1-1B is the photograph of negative control cells. In this figure we observed that there were no remarkable colored colonies. Fig2-2-4-1-2 shows the difference between cells storing P(3HB) and those not storing P(3HB) on one plate. The cells in blue rectangle area are the cells with P(3HB) synthesis gene and the cells in green rectangle area are the cells with PlasI-gfp gene as a negative control. Using the cells storing P(3HB), we drew a rose silhouette on the LB agar plate containing Nile red (Fig2-2-4-1-3).[Protocol]
4-2 Confirmation of P(3HB) accumulated in cells
To confirm the accumulation condition of P(3HB) in E.coli with a microscope, we stained the P(3HB) with Nile blue A reagent. Nile blue A is also used to detect the existence of P(3HB) and has no toxicity to the cells([5]). Before the observation, we stained the dried cells with Nile blue A solution. We then took photographs of the sample under fluorescence microscope. Fig2-2-4-2-1 is the photograph of dried E.coli (with pha C1-A-B1 gene) cells dyed with Nile blue A solution taken by fluorescence microscope. The fluorescent areas in Fig2-2-4-2-1A are the accumulated P(3HB) in the cells. This result also indicates that part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] synthesized P(3HB). In the photograph of negative control (Fig2-2-4-2-1B), no remarkable fluorescent area was observed.[Protocol]
4-3 Confirmation of P(3HB) by GC/MS
We successfully identified the products by [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] as 3HB, monomer of P(3HB), by Gas Chromatography/ Mass Spectrometry (GC/ MS). To confirm the products using GC/ MS, the products are methylated because 3HB is difficult to measure. Fig. 2-2-4-3-1 shows the GC/ MS result of the products by [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]. The peaks of sample are same to those of standard control of methylated 3HB. This shows that E.coli synthesized P(3HB) correctly. [Protocol]
4-4 Optimization of the best culture condition to synthesize P(3HB)
To figure out best culture condition, we tried culturing E.coli JM109 in 10 different conditions for 48h. Each condition is shown in Fig.2-2-4-4-1. Composition of LB and TB medium is shown in Fig. 2-2-4-4-2.
Pantothenic acid (PA), also called vitamin B5 is required to synthesize coenzyme A (CoA). If the glycolytic pathway has become a rate-limiting step, P(3HB) synthesis would be more efficiently by adding PA.
The culture result is shown in Fig. 2-2-4-4-4.
- “Dried cells (g/L)” is the amount of the cells in the medium after culturing.
- “Polymer content rate (%)” is the rate of the polymer in the dried cells.
- “Polymer concentration (g/L)” is the amount of the polymer in the medium after culturing. This value is calculated by multiplying “Dried cells” and “Polymer content rate”.
The results showed that TB medium was much better than LB medium to synthesize P(3HB). In both LB and TB, in the 37°C culturing containing glucose and PA-Ca, E.coli synthesized the polymer in maximum content rate. However, the growth of E.coli in 37°C was worse than that in 30°C, therefore final polymer concentration in 37°C and 30°C didn’t make a significant difference. Even if there was no glucose, E.coli synthesized polymer (condition 9 & 10). We think that TB medium had glycerol and a lot of yeast extra, and then E.coli might have used them as carbon sources. In addition, the comparison of condition 4 & 5 indicates PA-Ca was not used as carbon sources. LB medium didn’t contain many carbon sources, so E.coli synthesized little polymer. In this case, adding PA-Ca didn’t have big effect. On the other hand TB medium contains enough carbon sources, so we think that the rate-limiting step was the glycolytic pathway. In this case, polymer production would be increased by adding PA-Ca. (the comparison of condition 7 & 8 and 9&10)
[Protocol]
Application
The achievement of our project “P(3HB) Production” is that we registered available P(3HB) synthetic gene in Biobrick parts. We can control the expression of the P(3HB) synthetic gene spatially by using combination of Biobrick parts. What we want to claim as an example of the spatial manipulation of gene expression is water-repellent. A stronger water-repellent requires hydrophobicity as well as the increase in real surface area that can be achieved as ruggedness of P(3HB) adsorbed on particular surface. If we can control the expression of the P(3HB) synthetic gene spatially by using genetic parts which are registered in Biobrick parts, the application of a super water-repellent sheet will become available.
We made P(3HB) sheets. Contact angle is an indicator to represent the strength of the water-repellent. The angle shows the physical properties, especially surface tension.When contact angle of sheets is larger than 90°, from Young equation, the sheets would have stronger water-repellent by increasing real surface area. From literature data, contact angle of P(3HB) sheets is about 100°.[Protocol]
Reference
[1] Jumiarti Agus, Altered expression of polyhydroxyalkanoate synthase gene and its effect on poly[(R)-3-hydroxybutyrate] synthesis in recombinant Escherichia coli, Polymer Degradation and Stability(2006) 91:1645-1650
[2] Joanne Stubbe and Jiamin Tian, Polyhydroxyalkanoate (PHA) homeostasis: the role of the PHA synthase, 2003, Nat. Prod. Rep.,20, 445–457.
[3] Stanley D. Fowler and Phillip Greenspan, Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections, Histochemistry & Cytochemistry(1985), vol 33.No 8, 833-836
[4] Pinzon NM, Nile red detection of bacterial hydrocarbons and ketones in a high-throughput format, mBio (2011),vol 2. issue 4.e-00109-11
[5] Patricia Spiekermann, A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds, Arch Microbiol (1999), 171:73–80
[6] Vladimir K. Vanag, Cross-diffusion and pattern formation in reaction–diffusion systems, Physical Chemistry Chemical Physics(2009), vol 11.897-912
[7] Pohlmann A, et al, Genome sequence of the bioplastic-producing "Knallgas" bacterium Ralstonia eutropha H16, Nat Biotechnol(2006), 24:1257-62 "
"