Team:Tokyo Tech/Experiment/PHB
From 2012.igem.org
(→D. Optimization of the best culture condition to synthesize P(3HB)) |
(→D. Optimization of the best culture condition to synthesize P(3HB)) |
||
| Line 75: | Line 75: | ||
The results show that TB medium is much better than LB medium to synthesize P(3HB). In both LB and TB, in the 37°C culturing containing glucose and PA-Ca, <I>E.coli</I> synthesizes the polymer in maximum content rate. However, the growth of <I>E.coli</I> in 37°C is worse than that in 30°C, therefore final polymer concentration in 37°C and 30°C doesn’t make a significant difference. Even if there is no glucose, <I>E.coli</I> synthesizes polymer (condition 9 & 10). We think that TB medium has glycerol and a lot of yeast extra, then <I>E.coli</I> may use them as carbon sources. | The results show that TB medium is much better than LB medium to synthesize P(3HB). In both LB and TB, in the 37°C culturing containing glucose and PA-Ca, <I>E.coli</I> synthesizes the polymer in maximum content rate. However, the growth of <I>E.coli</I> in 37°C is worse than that in 30°C, therefore final polymer concentration in 37°C and 30°C doesn’t make a significant difference. Even if there is no glucose, <I>E.coli</I> synthesizes polymer (condition 9 & 10). We think that TB medium has glycerol and a lot of yeast extra, then <I>E.coli</I> may use them as carbon sources. | ||
| - | In addition, the comparison of condition 4 & 5 indicates PA-Ca is not used as carbon sources. LB medium doesn’t contain many carbon sources, so <I>E.coli</I> synthesizes little polymer. In this case, adding PA-Ca doesn’t have big effect. On the other hand TB medium contains enough carbon sources, so we think that the rate-limiting step is the glycolytic pathway. In this case, polymer production would be increased by adding PA-Ca.(the comparison of condition 7 & 8 and 9 & 10)[[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#D. | + | In addition, the comparison of condition 4 & 5 indicates PA-Ca is not used as carbon sources. LB medium doesn’t contain many carbon sources, so <I>E.coli</I> synthesizes little polymer. In this case, adding PA-Ca doesn’t have big effect. On the other hand TB medium contains enough carbon sources, so we think that the rate-limiting step is the glycolytic pathway. In this case, polymer production would be increased by adding PA-Ca.(the comparison of condition 7 & 8 and 9 & 10)[[https://2012.igem.org/Team:Tokyo_Tech/Experiment/PHB#D._Optimization_of_the_best_culture_condition_to_synthesize_P.283HB.29. Protocol]] |
=Construction of pha-C1-A-B1 in Biobrick format= | =Construction of pha-C1-A-B1 in Biobrick format= | ||
Revision as of 02:49, 27 October 2012
Contents |
P(3HB) production by E.coli & Confirmation of P(3HB)
To synthesize P(3HB) by E.coli, we transformed E.coli JM109 with the constructed pha C1-A-B1 part on pSB1C3 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]). E.coli JM109 is used to synthesize P(3HB), because it tends to have a high density accumulation of P(3HB)([5] ). As a negative control, we transformed E.coli JM109 with PlasI-gfp on pSB1C3.
A. Confirmation of P(3HB) synthesized on colonies
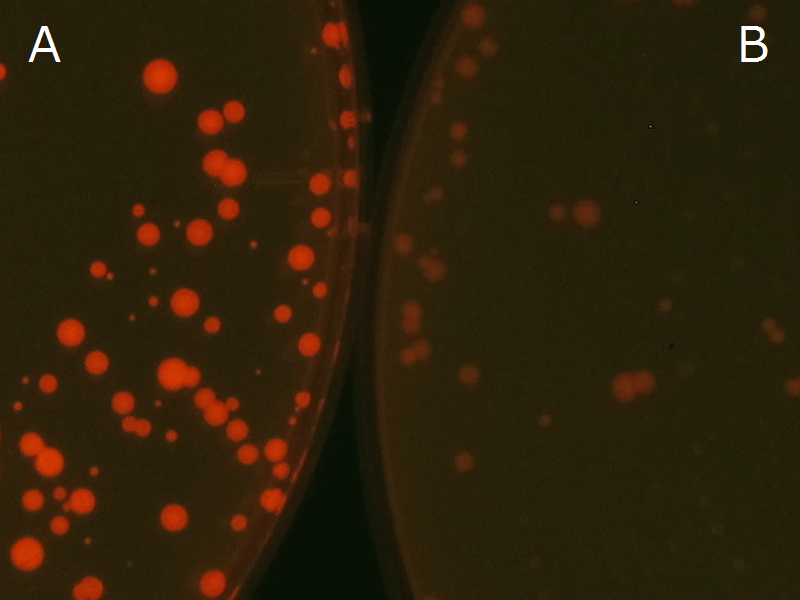
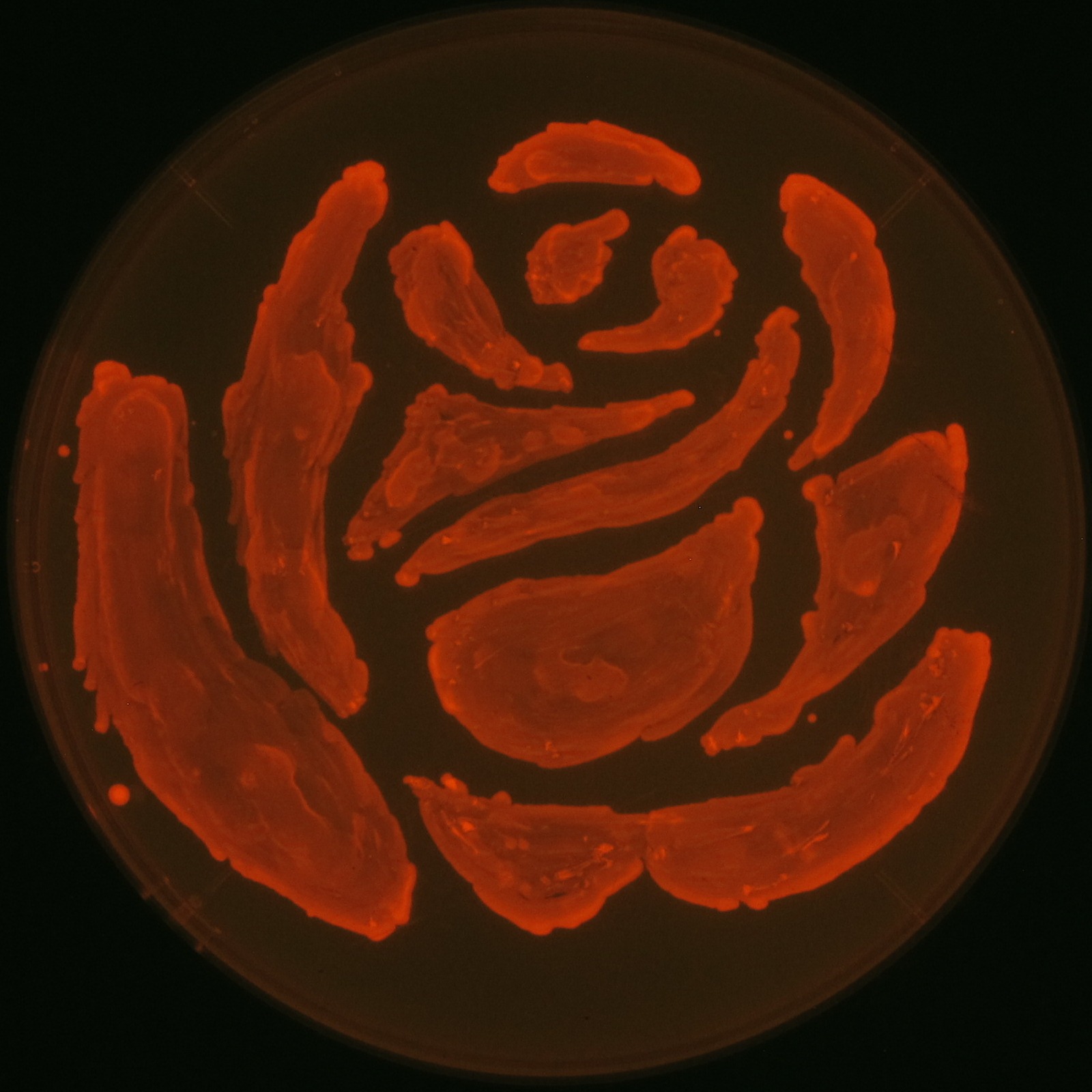
We observed the accumulation of P(3HB) in the E.coli colonies on Nile red positive medium under UV. Nile red has been widely used to stain colonies and distinguish between PHA-accumulating and non-accumulating colonies. Nile red in the agar medium doesn’t affect the growth of the cells, and the accumulation of PHAs in the colonies can be directly monitored([3][4][5] ). We cultured the transformant on LB agar medium plates with Nile red. After several days, colonies storing P(3HB) were stained orange by Nile red when observed under UV. This result indicates that transformant synthesized and stored P(3HB). Fig2-2-4-1-1 is the photographs of E.coli colonies on Nile red positive medium taken under UV. The orange colonies in Fig2-2-4-1-1A show that the accumulated P(3HB) in cells was stained by Nile red. This result indicates that part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] synthesized P(3HB). Fig2-2-4-1-1B is the photograph of negative control cells. In this figure we observed that there were no remarkable colored colonies. Fig2-2-4-1-2 shows the difference between cells storing P(3HB) and those not storing P(3HB) on one plate. The cells in blue rectangle area are the cells with P(3HB) synthesis gene and the cells in green rectangle area are the cells with PlasI-gfp gene as a negative control. Using the cells storing P(3HB), we drew a rose silhouette on the LB agar plate containing Nile red (Fig2-2-4-1-3).[Protocol]
B. Confirmation of P(3HB) accumulated in cells
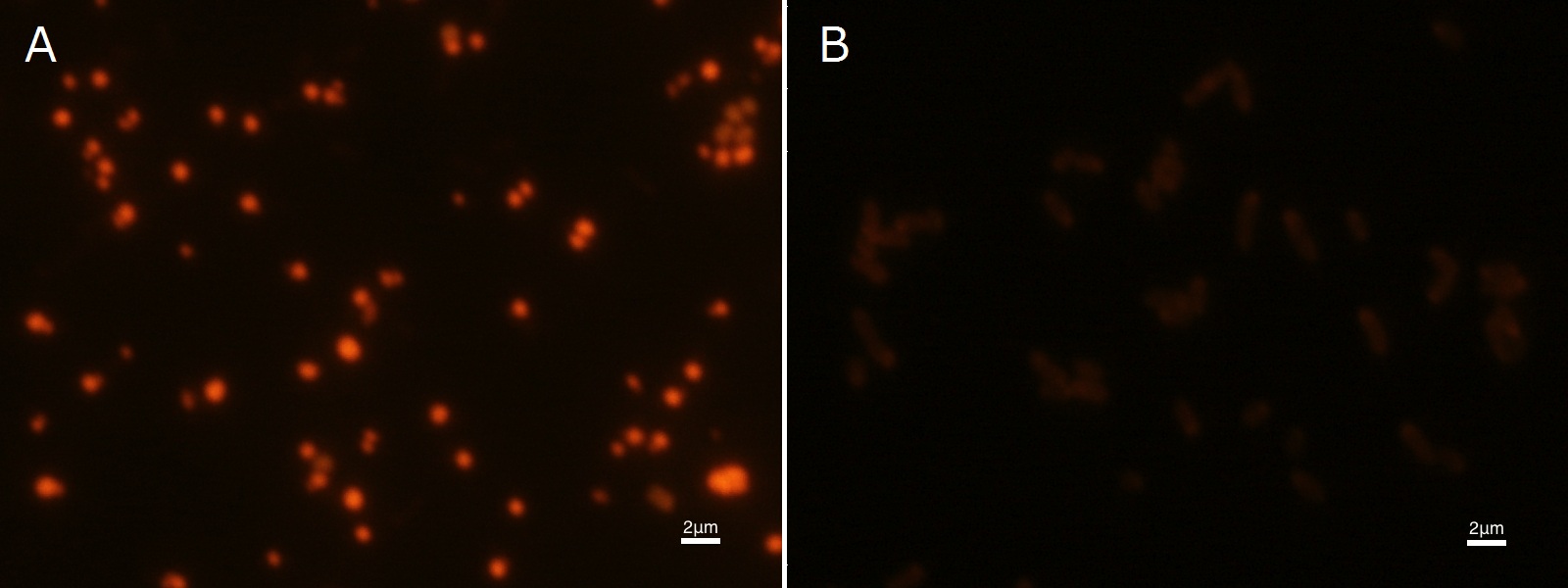
To confirm the accumulation condition of P(3HB) in E.coli with a microscope, we stained the P(3HB) with Nile blue A reagent. Nile blue A is also used to detect the existence of P(3HB) and has no toxicity to the cells([5]). Before the observation, we stained the dried cells with Nile blue A solution. We then took photographs of the sample under fluorescence microscope. Fig2-2-4-2-1 is the photograph of dried E.coli (with pha C1-A-B1 gene) cells dyed with Nile blue A solution taken by fluorescence microscope. The fluorescent areas in Fig2-2-4-2-1A are the accumulated P(3HB) in the cells. This result also indicates that part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] synthesized P(3HB). In the photograph of negative control (Fig2-2-4-2-1B), no remarkable fluorescent area was observed.[Protocol]
C. Confirmation of P(3HB) by GC/MS
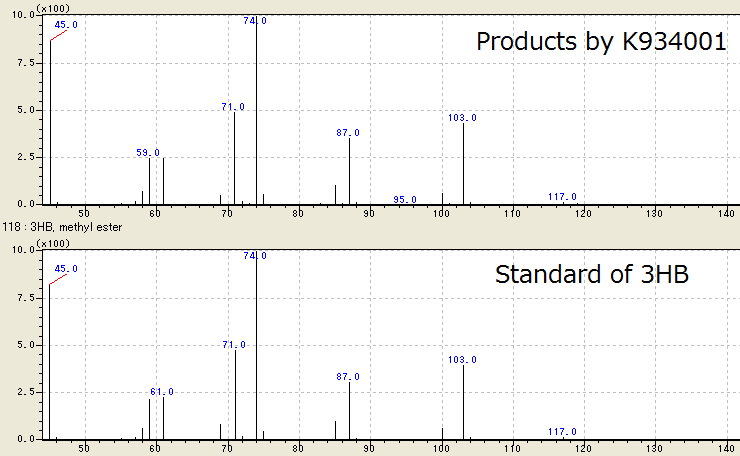
We successfully identified the products by [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] as 3HB, monomer of P(3HB), by Gas Chromatography/ Mass Spectrometry (GC/ MS). To confirm the products using GC/ MS, the products are methylated because 3HB is difficult to measure. Fig. 2-2-4-3-1 shows the GC/ MS result of the products by [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]. The peaks of sample are same to those of standard control of methylated 3HB. This shows that E.coli synthesized P(3HB) correctly. [Protocol]
D. Optimization of the best culture condition to synthesize P(3HB)
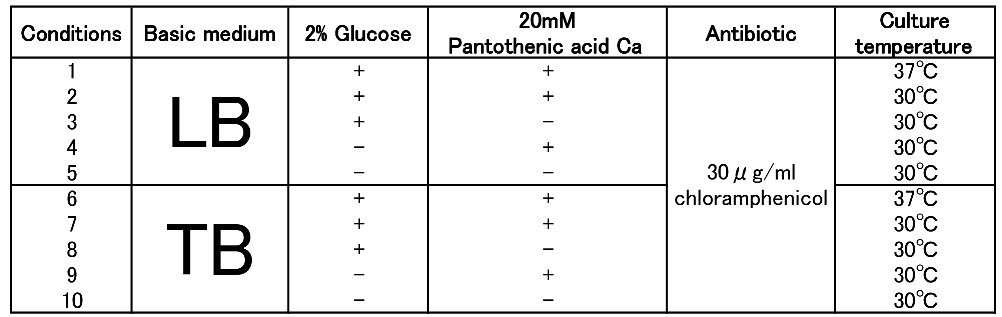
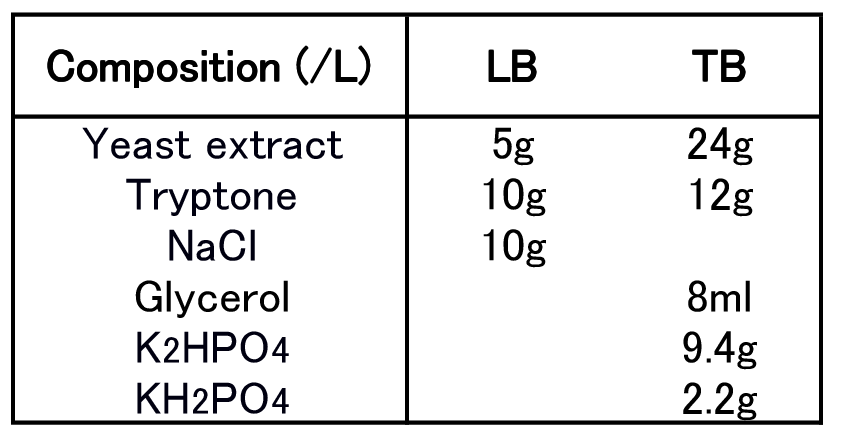
To figure out best culture condition, we tried culturing E.coli JM109 in 10 different conditions for 48h. Each condition is shown in Fig.2-2-4-4-1. Composition of LB and TB medium is shown in Fig. 2-2-4-4-2.
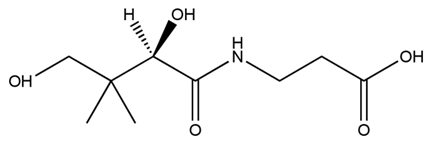
Pantothenic acid (PA), also called vitamin B5 is required to synthesize coenzyme A (CoA). If the glycolytic pathway has become a rate-limiting step, P(3HB) synthesis would be more efficiently by adding PA.
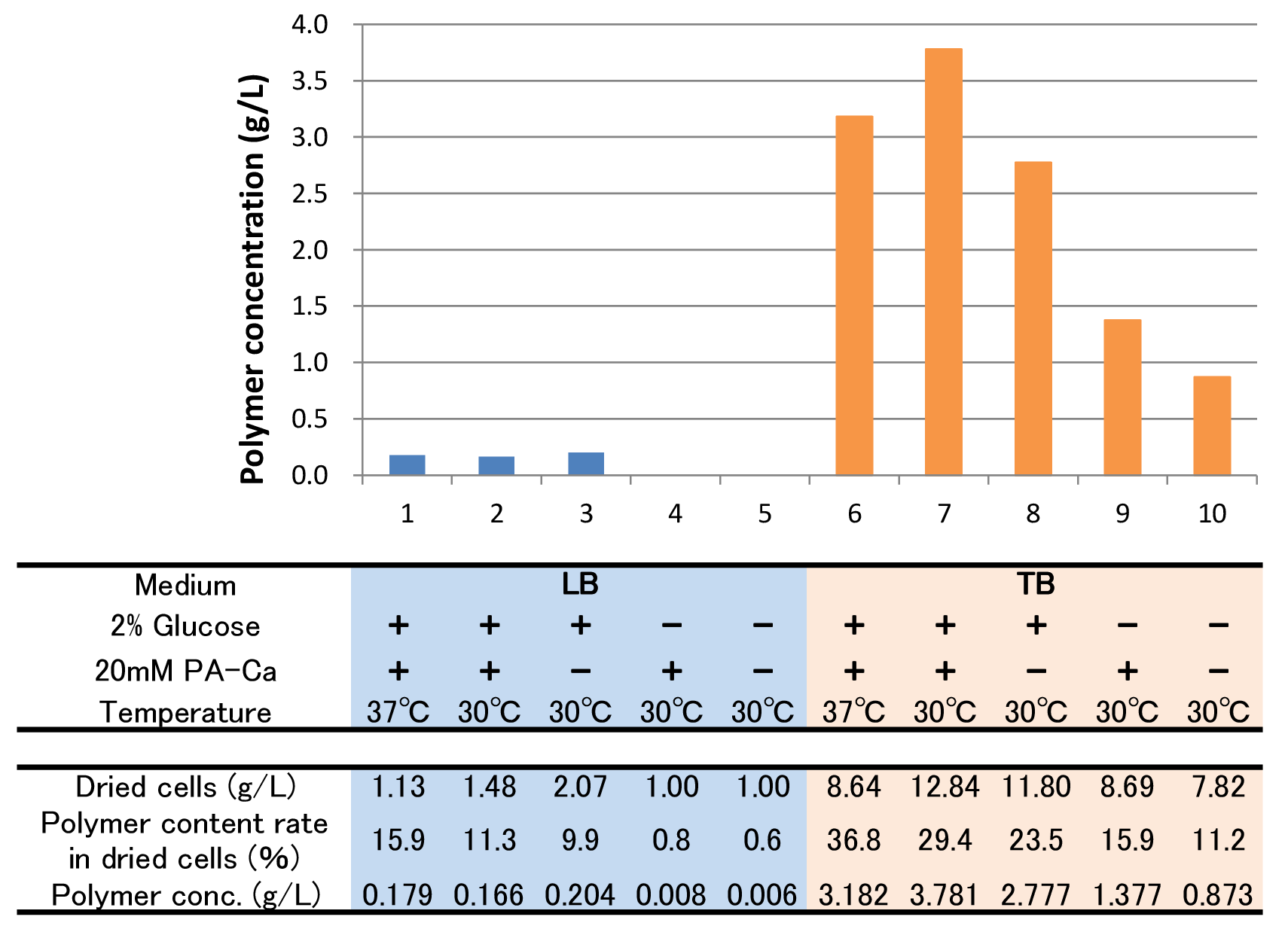
The culture result is shown in Fig. 2-2-4-4-4.
- “Dried cells (g/L)” is the amount of the cells in the medium after culturing.
- “Polymer content rate (%)” is the rate of the polymer in the dried cells.
- “Polymer concentration (g/L)” is the amount of the polymer in the medium after culturing. This value is calculated by multiplying “Dried cells” and “Polymer content rate”.
The results show that TB medium is much better than LB medium to synthesize P(3HB). In both LB and TB, in the 37°C culturing containing glucose and PA-Ca, E.coli synthesizes the polymer in maximum content rate. However, the growth of E.coli in 37°C is worse than that in 30°C, therefore final polymer concentration in 37°C and 30°C doesn’t make a significant difference. Even if there is no glucose, E.coli synthesizes polymer (condition 9 & 10). We think that TB medium has glycerol and a lot of yeast extra, then E.coli may use them as carbon sources.
In addition, the comparison of condition 4 & 5 indicates PA-Ca is not used as carbon sources. LB medium doesn’t contain many carbon sources, so E.coli synthesizes little polymer. In this case, adding PA-Ca doesn’t have big effect. On the other hand TB medium contains enough carbon sources, so we think that the rate-limiting step is the glycolytic pathway. In this case, polymer production would be increased by adding PA-Ca.(the comparison of condition 7 & 8 and 9 & 10)[Protocol]
Construction of pha-C1-A-B1 in Biobrick format
[Back to "Construction of phaC1-A-B1 in Biobrick format"]
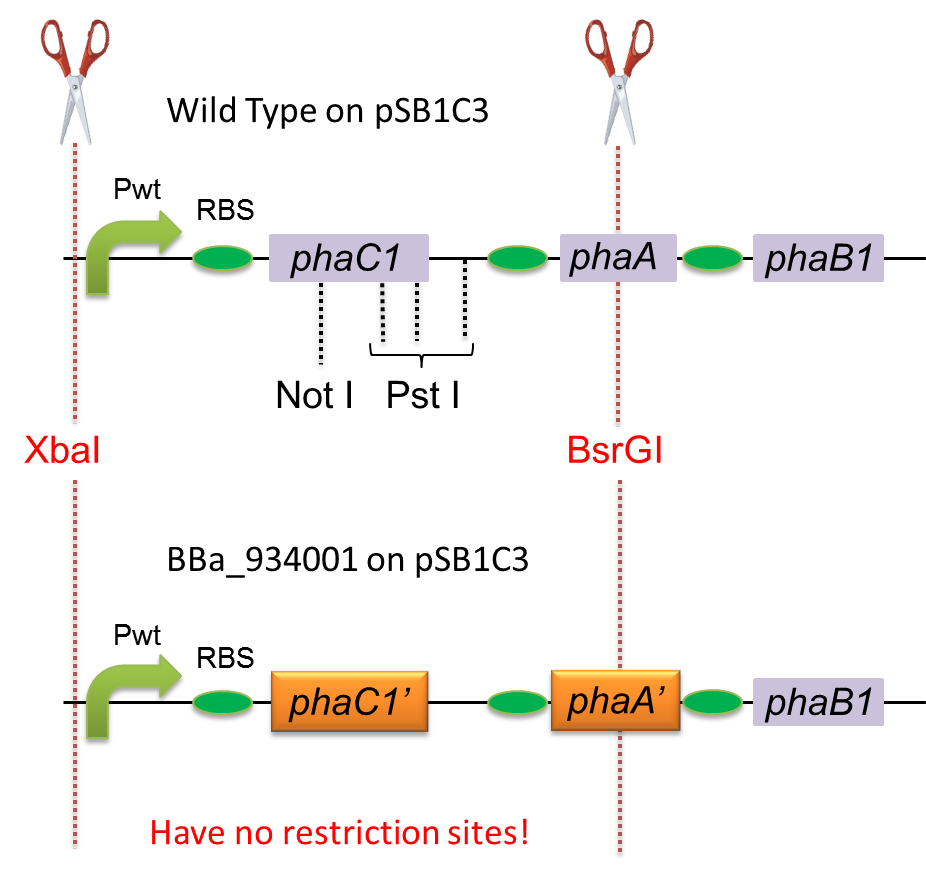
To construct a part that meets Biobrick format, we have modified the phaC1-A-B1 operon not to contain forbidden restriction enzyme sites. First, we cloned the wild type gene phaC1-A-B1 from R.eutropha H16 by using PCR and inserted the gene into pSB1C3. However, wild type phaC1-A-B1 gene sequence contains one NotI and three PstI recognition sites that are not allowed in Biobrick format. To get phaC1-A-B1 sequence without these recognition sites, we ordered the chemically synthesized DNA from IDT/MBL. In this chemically synthesized DNA, coding is optimized for E.coli. We used restriction enzyme XbaI (on pSB1C3) and BsrGI (on phaC1-A-B1) to insert sequence. That is to say, we got Poly[(R)-3-hydroxybutyrate] synthesizing gene in Biobrick format ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]).
[Back to "Construction of phaC1-A-B1 in Biobrick format"]
Protocol
A .P(3HB) production on colonies and preparation before confirmation with Nile red under UV
[Back to "4-1 Confirmation of P(3HB) synthesized on colonies"]
1 Preparation of LB agar medium plate containing Nile red and Glucose
1.1 Autoclave a LB agar(final 40g/L) solution at 120 ° C
1.2 After the autoclave, add Chloramphenicol(final 25ug/ml), Nile red and glucose(final 20g/L) to the LB agar solution when it cools down.
1.3 Make LB agar medium plates with the mixture.
2 Transformation of E.coli strain JM109 with pSB1C3 plasmid containing phaC1-A-B1 into strain JM109
2.1 Thaw the competent cells JM109 at 4° C
2.2 Add the target DNA 3ul into 1.5ml tube, then add in 50ul the thawed competent cells.
2.3 Put the tube into ice for 15mins
2.4 42° C,30secs, heatshock
2.5 Add 160ul of SOC into the tube
2.6 Incubate the the cells at 37° C for 30mins
2.7 Spread the resulting culture on LB agar medium plate with a large cone rod.
2.8 Incubate the plate at 37° C for 36hrs then cells the plate into 4° C room for 2-3 days.
[Back to "4-1 Confirmation of P(3HB) synthesized on colonies"]
B.P(3HB) production in cells and preparation before the confirmation with Nile blue A
[Back to "4-2 Confirmation of P(3HB) accumulated in cells"]
1 Production of PHB
1.1 Acquire one colony of the transformed strains (JM109) with a platinum loop
1.2 Culture the colony in LB solution for 16hrs at 37 ° C
1.3 Measure LB medium (final 2.5%) and add it to each Erlenmeyer flask inside clean bench.
1.4 Add distilled water(final 95%) to each Erlenmeyer flask and cover the flasks with four-folded aluminum foil.
1.5 Set all flasks into autoclave
1.6 Add Chloramphenicol(final 25ug/ml) and glucose solution (50%) (final 20g/L) after the medium is completely cooled.
1.7 Add the solution of cultured cells into each flasks and shaking culture with air permeable lids at 37 ° C for 96 hours.
2 Preparation before the confirmation (with Nile blue A) under fluorescent microscope
2.1 Collection of PHBs in JM109
2.1.1 Weigh empty 50ml falcon tube without lid and make a record.
2.1.2 Add some culture solution into each tube.
2.1.3 Set the tubes into centrifuge and make sure that the label faces outside.
2.1.4 4 ° C, 5000G, 10mins in centrifuge.
2.1.5 Remove the supernatant with electric pipettor then add culture solution and set in centrifuge again.
2.1.6 After adding all the culture solution and setting in centrifuge, remove the supernatant and add water, set in centrifuge again.
2.1.7 Remove the supernatant and add a little amount of water
2.1.8 Cover the tubes with double layers of parafilms and fully freeze them.
2.2 Freeze drying (lyophilization)
2.2.1 Poke several holes on the tubes’ parafilm with toothpick.
2.2.2 Set the tubes on the freeze drying machine.
2.2.3 Freeze dry for 3 days.
2.3 Stain PHB accumulated dried cells with Nile blue A before observation
2.3.1 Acquire dried cells after freeze drying
2.3.2 Put a small amount of cells on the slide glass
2.3.3 Add water on the cells and heat the slide glass immobilize the cells
2.3.4 Stain the cells with 1% Nile blue A solution (water) for 8 minutes
2.3.5 Wash excess Nile blue A with 8% acetic acid solution
C. Preparation for GC/MS
[Back to "4-3 Confirmation of P(3HB) by GC/MS"]
1. Put 10mg of dried cells in glass tubes.
2. Add 2ml MeOH (containing 15% sulfuric acid) and 2ml chloroform.
3. Incubate tubes at 100℃ for 140min.
4. Add 1ml pure water, stir tubes, and incubate until the solution became clear.
5. Remove the organic layer, filtered.
6. Add internal standard fluid to organic layer.
7. Set in GC/MS.
D. Optimization of the best culture condition to synthesize P(3HB).
[Back to "Optimization best culture condition to synthesize P(3HB)"]
1 Preparing
1.1 2x LB solution (autoclaved) 100ml
Tryptone 2g
Yeast extract 1g
NaCl 2g
1.2 2x TB solution (autoclaved) 100ml
Tryptone 2.4g
Yeast extract 4.8g
Glycerol 1.6ml
K2HPO4 1.88g
KH2PO4 0.44g
1.3 50% glucose (autoclaved) 100ml
Glucose 50g
Pure water up to 100ml
1.4 1M Pantothenic acid Ca (Filter sterilized)
Pantothenic acid Ca 9.53g
Pure water up to 20ml
2 Polymer producing media
2.1 LB, 2% Glc, 20mM Pantothenic acid Ca, 30μg/ml Chloramphenicol, 10ml
2x LB 5ml
50% Glc 400ul
1M Pantothenic acid Ca 200ul
Cm(25mg/ml) 12ul
Pure water(autoclaved) 4.388ml
2.2 TB, 2% Glc, 20mM Pantothenic acid Ca, 30μg/ml Chloramphenicol, 10ml
2x TB 5ml
50% Glc 400ul
1M Pantothenic acid Ca 200ul
Cm(25mg/ml) 12ul
Pure water(autoclaved) 4.388ml
2.3 LB, 2% Glc, 30μg/ml Chloramphenicol, 10ml
2x LB 5ml
50% Glc 400ul
Cm(25mg/ml) 12ul
Pure water(autoclaved) 4.588ml
2.4 TB, 2% Glc, 30μg/ml Chloramphenicol, 10ml
2x TB 5ml
50% Glc 400ul
Cm(25mg/ml) 12ul
Pure water(autoclaved) 4.588ml
2.5 LB, 20mM Pantothenic acid Ca, 30μg/ml Chloramphenicol, 10ml
2x LB 5ml
Pantothenic acid Ca 200ul
Cm(25mg/ml) 12ul
Pure water(autoclaved) 4.788ml
2.6 TB, 20mM Pantothenic acid Ca, 30μg/ml Chloramphenicol, 10ml
2x TB 5ml
Pantothenic acid Ca 200ul
Cm(25mg/ml) 12ul
Pure water(autoclaved) 4.788ml
2.7 LB, 30μg/ml Chloramphenicol, 10ml
2x LB 5ml
Cm(25mg/ml) 12ul
Pure water(autoclaved) 4.988ml
2.8 TB, 30μg/ml Chloramphenicol, 10ml
2x TB 5ml
Cm(25mg/ml) 12ul
Pure water(autoclaved) 4.988ml
3. Culture and collection
3.1 Use LB medium to preculture transformed media 1.5 ml for 15 hrs, 180 rpm/ 37℃.
3.2 Culture 15 μl preculture media into different conditions for 48 hrs, 180 rpm.
3.3 Collect cells and centrifuge for 3 min, 5,000 rpm.
3.4 Remove supernatant and suspend with pure water.
3.5 Centrifuge again for 3 min, 5,000 rpm and remove its supernatant.
3.6 Freeze in -20℃.
3.7 Freeze-dry for 72 hrs.
[Back to "Optimization best culture condition to synthesize P(3HB)"]
Reference
[1] Jumiarti Agus, Altered expression of polyhydroxyalkanoate synthase gene and its effect on poly[(R)-3-hydroxybutyrate] synthesis in recombinant Escherichia coli, Polymer Degradation and Stability(2006) 91:1645-1650
[2] Joanne Stubbe and Jiamin Tian, Polyhydroxyalkanoate (PHA) homeostasis: the role of the PHA synthase, 2003, Nat. Prod. Rep.,20, 445–457.
[3] Stanley D. Fowler and Phillip Greenspan, Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections, Histochemistry & Cytochemistry(1985), vol 33.No 8, 833-836
[4] Pinzon NM, Nile red detection of bacterial hydrocarbons and ketones in a high-throughput format, mBio (2011),vol 2. issue 4.e-00109-11
[5] Patricia Spiekermann, A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds, Arch Microbiol (1999), 171:73–80
[6] Vladimir K. Vanag, Cross-diffusion and pattern formation in reaction–diffusion systems, Physical Chemistry Chemical Physics(2009), vol 11.897-912
[7] Pohlmann A, et al, Genome sequence of the bioplastic-producing "Knallgas" bacterium Ralstonia eutropha H16, Nat Biotechnol 24:1257-62 (2006) "
"