Team:Tokyo Tech/Experiment/PHB
From 2012.igem.org
(Created page with " =Construction of pha-C1-A-B1 in Biobrick format= [[https://2012.igem.org/Team:Tokyo_Tech/Projects/PHAs/index.htm#Construction_of_phaC1-A-B1_in_Biobrick_format Back to "Construct...") |
|||
| Line 1: | Line 1: | ||
| + | {{tokyotechcss}} | ||
| + | {{tokyotechmenubar}} | ||
| + | <br><br> | ||
| + | |||
| + | <div class="whitebox"> | ||
| + | =PHB production= | ||
Revision as of 04:26, 17 October 2012
PHB production
Construction of pha-C1-A-B1 in Biobrick format
[Back to "Construction of phaC1-A-B1 in Biobrick format"]
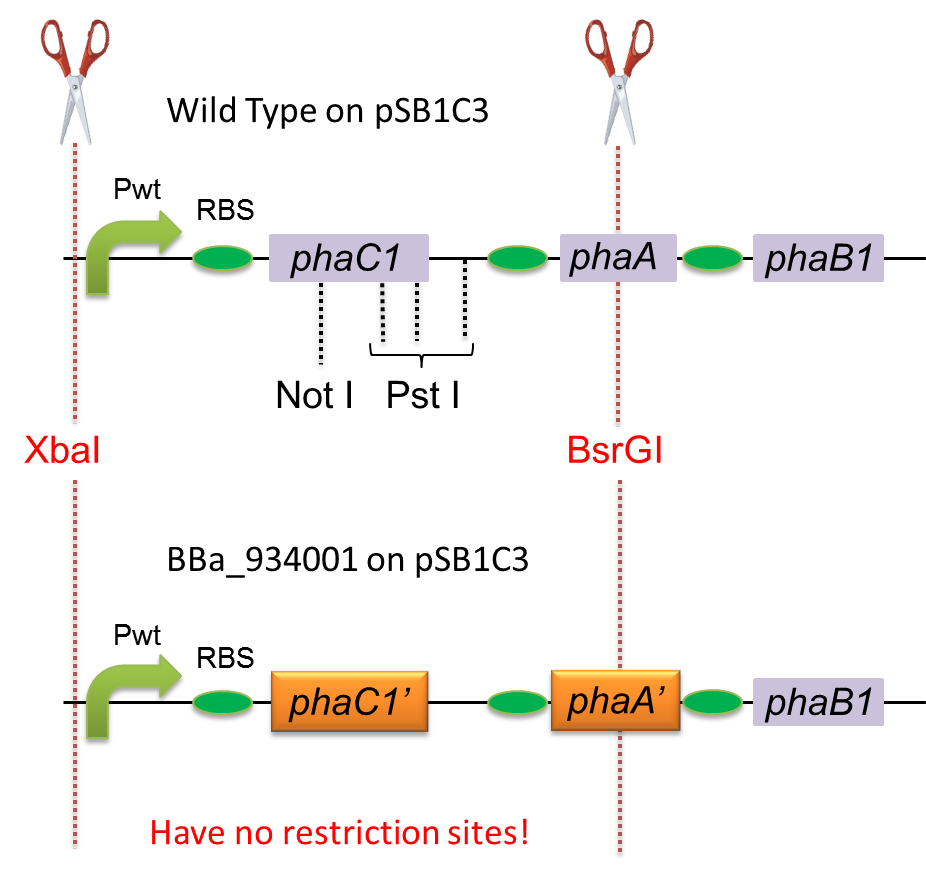
To construct a part that meets Biobrick format, we have modified the phaC1-A-B1 operon not to contain forbidden restriction enzyme sites. First, we cloned the wild type gene phaC1-A-B1 from R.eutropha H16 by using PCR and inserted the gene into pSB1C3. However, wild type phaC1-A-B1 gene sequence contains one NotI and three PstI recognition sites that are not allowed in Biobrick format. To get phaC1-A-B1 sequence without these recognition sites, we ordered the chemically synthesized DNA from IDT/MBL. In this chemically synthesized DNA, coding is optimized for E.coli. We used restriction enzyme XbaI (on pSB1C3) and BsrGI (on phaC1-A-B1) to insert sequence. That is to say, we got Poly[(R)-3-hydroxybutyrate] synthesizing gene in Biobrick format (BBa_K934001).
[Back to "Construction of phaC1-A-B1 in Biobrick format"] "
"