Team:Peking/Project/Phototaxis
From 2012.igem.org
Introduction
Bacterial phototaxis refers to the ability of bacteria to sense light changes in their extracellular environment and to bias their motility towards or away from the light. Phototactic responses are observed in many organisms such as Serratia marcescens, Tetrahymena, and Euglena. The behavior of phototaxis is the directed movement up a gradient to an increasing amount of light. This is analogous to positive chemotaxis -- the usage of light as the attractant rather than chemicals.
Figure 1. Phototactic behavior of bacteria
For synthetic biology, phototaxis is a palace of mysteries because it has a rapid, precise response in comparison with chemotaxis while its mechanism has not been fully elucidated so far. As chemicals diffuse in the culture medium, chemotaxis does not have quite enough precision as many would prefer. On the contrary, light is noninvasive, reversible and of good spatiotemparol control. With these advantages, phototaxis is more qualified for the application than chemotaxis in synthetic biology. Also, light signal is a potential link between the electrical components and bio-elements that endows phototaxis with a great potential of technology improvement such as light-guided cell targeting for medicine aggregation in the body or tissue recovery in biomedical applications, light-induced bacterial enrichment or dispersion in environment protection and so forth. (Figure 1)
The model of chemotaxis pathway is quite clear and widely accepted. E.coli alternates between a running mode(smooth swimming) and a tumbling mode, which results from distinct types of flagellar rotation, counter-clockwise(CCW) and clockwise(CW), respectively. A number of intracellular proteins provide the necessary signaling cascade which links the membrane receptors to the flagellar: CheY, CheZ, CheW, CheA, (Figure 2) etc. (See Modeling Phototaxis)
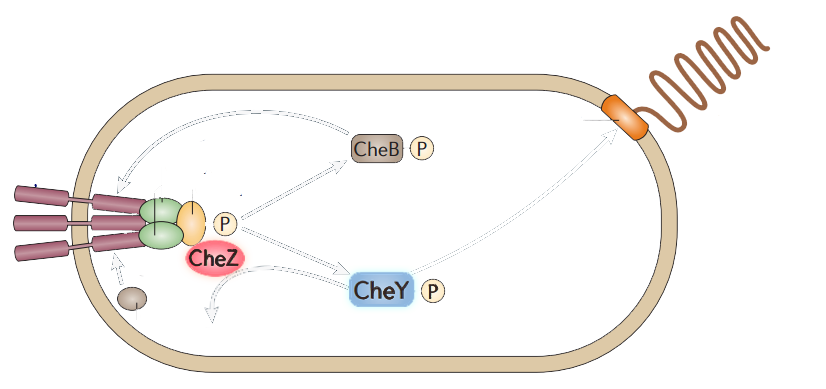
Figure 2. The simplified scheme of protein-protein interactions during chemotaxis of bacteria.
Among those proteins, CheY have the direct attachment to the flagella motors. Phosphorylated CheY diffuses through the cytoplasm to the motors,binding to FliM, a component of the C ring of the flagellar motor, and induces CW rotation of the motor. CheZ can dephosphorylate CheY, resulting in CCW rotations which leads to the smooth swimming. CheZ mutants result into swarming while CheY mutants are smooth-swimming.
CheZ is commonly used to affect bacterial motion in synthetic biology. Experiments shows that different levels of CheZ induced by arabinose can change the diameter of the swarming colony, combination between the quorum sensing part and CheZ can make bacteria form a stripe pattern. So we suppose combination between CheZ and our Luminesensor must have an effect on bacteria in motion. As modeling reveals, it does have a phototaxis phenomenon where light is a negative stimuli, showing that it is very promising to build a ultra-sensitive, effective, promising phototaxis design.
Reference
- 1. Kuo, S.C., and Koshland, D.E.(1987) Roles of che Y and cheZ Gene Products in Controlling Flagellar Rotation in Bacterial Chemotaxis of Escherichia coli. J. Bacteriol., 3:1307:1313
- 2. Bren, A., Welch, M., Blat, Y., Eisenbeach, M.(1996) Signal termination in bacterial chemotaxis: CheZ mediates dephosphorylation of free rather than switch-bound CheY. Proc. Natl. Acad. Sci. USA, 93: 10090: 10093
- 3. Bren, A., and Eisenbeach, M.(2000) How Signals Are Heard during Bacterial Chemotaxis: Protein-Protein Interactions in Sensory Signal Propagation. J. Bacteriol., 182: 6865: 6873
- 4. Sanna, M. G., and Simon, M.I.(1996) in vivo and in vitro Characterization of Escherichia coli Protein CheZ Gain- and Loss-of-Function Mutants. J. Bacteriol., 178: 6275: 6280
- 5. Liu, C., et al.(2012) Sequential Establishment of Stripe Patterns in an Expanding Cell Population. Science, 334: 238: 241
- 6. Lee, S.H., Butler, S.M., and Camilli, A.(2001) Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98: 6889: 6894
- 7. sinha, J., Reyes, S.J., Gallivan, J.P.(2010) Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol., 464:468
- 8. Topp, S., and Gallivan. J.P.(2008) Random Walks to Synthetic Riboswitches — A High-Throughput Selection Based on Cell Motility. Chem. Biol., 9:210:213
 "
"














