Team:Peking/Project/Overview
From 2012.igem.org
| Line 104: | Line 104: | ||
</p> | </p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="" alt="" style="width:500px;" /> | + | <img src="/wiki/images/c/c1/Peking2012_Home_Phototaxis.jpg" alt="" style="width:500px;" /> |
<div> | <div> | ||
<p class="description"> | <p class="description"> | ||
Revision as of 17:04, 23 September 2012
Achievements
Here is a list of our achievements from this summer.
1. We helped four other iGEM teams by sharing DNA materials, characterizing their parts and modeling! Click Here
2. We outlined in detail a new approach of human pratice called "Sowing Tomorrow's Synthetic Biologists"! Click Here
3. We presented all fresh iGEMers with a collection and praise of historic iGEM projects to share and learn from each other!Click Here
4. We submitted 11 high quality and well-characterized standard biobricks! Click Here
5. We improved the ease-of-use of "Lux Brick" that primarily constructed by Cambridge iGEM 2010 and carefully characterized its dynamics of functionality! Click Here
6. We rationally constructed an ultra-sensitive genetically encoded sensor of luminance – Luminesensor and comprehensively characterized it. Luminesensor proved to be as sensitive to sense natural light and even bioluminescence! Click Here
7. We successfully implemented spatiotemporal control of cellular behavior, such as high-resolution 2-D and 3-D bio-printing using dim light, and even utilizing the luminescence of an iPad! Click Here
8. We successfully implemented cell-cell communication using light for the very first time in synthetic biology! Click Here
We deserve a Gold Medal Prize.
1. The Luminesensor
-- An Ultrasensitive Light Sensor
Based on comprehensive literature reviews, Peking iGEM has concluded that an ultra-sensitive light-sensing transcription factor may circumvent serious issues of current optogenetic methods, e.g. cytotoxicity, narrow dynamic range, and dependency on laser and exogenous chromophores.
Inspired by the general design principle of genetically encoded sensors in chemical biology, we fused a light-sensing domain, the smallest LOV (light, oxygen, or voltage) protein- Vivid (VVD) from Neurospora, to the DNA binding domain of bacterial LexA, a paradigm of bacterial repressors. When illuminated by blue light (440~480nm), the dimerization of VVD and the DNA binding domain of LexA will bind to DNA to repress the expression of downstream gene(s) (Figure 1).
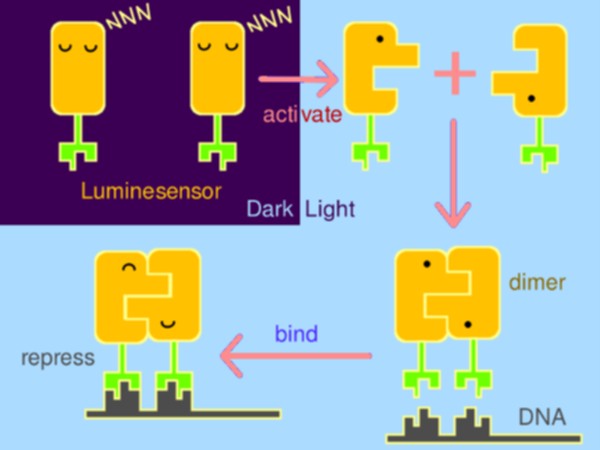
Figure 1. The basic design of the Luminesensor.
After primary construction of the fusion protein, protein structure inspection and kinetic simulations were conducted to rationally perform optimization. (If you would like to know details regarding the Luminesensor design, please see Design page.)
Due to the consideration of minimal interference with host genetic context, a special mutation at the LexA DNA binding domain was introduced to ensure orthogonality with the bacterial endogenous LexA protein. The resulting light-sensing protein proved to work very well in wild type E. coli strains without the necessity to genetically modify the host organism (data in Characterization).
Consistent with the results of kinetic simulation (details in Modeling), mutagenesis based on kinetic data of VVD mutation significantly improved the performance of our light-sensing protein, called the Luminesensor, now with an amazing dynamic range reaching 300 (data in Characterization).
The high sensitivity of the VVD protein is preserved in our Luminesensor. Amazingly, the Luminesensor proved to be sensitive to natural light and even bioluminescence (data in Characterization). Moreover, the Luminesensor is able to respond to light signals with broad illuminance scales (Figure 2).
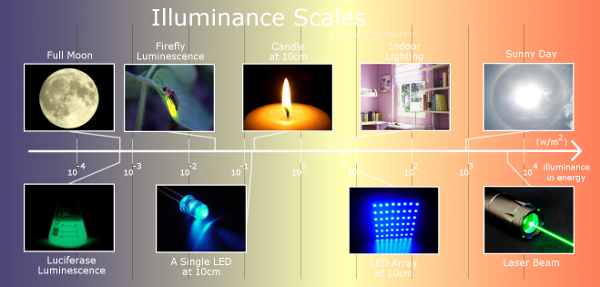
Figure 2. The broad illuminance scales which the Luminesensor could respond to.
To expand the toolkit of the Luminesensor, we also tried to red-shift the absorption spectrum of VVD through molecular docking simulations (see Modeling) and VVD mutation (see Extension), which would largely extend the future application of the Luminesensor, e.g. a multicolor Luminesensor control system.
2. Synthetic Biology in 2D and 3D
-- A Practical Application
Few optogenetics methods have been applied to practical applications until now. To fully demonstrate spatiotemporally precise manipulation of cellular behavior using our Luminesensor, Peking iGEM has deeply explored the applications in 2D and 3D bio-printing.
High resolution images could be easily obtained by 2D bioprinting on a bacterial lawn bearing the Luminesensor (Figure 3). (more on 2D Printing).

Figure 3. High resolution of 2D bioprinting.
What's more, sharp images could be printed using an Apple iPad as light source (see iPrinting), which indicates that our Luminesensor could serve as the interface between biological systems and electrical devices.
A very promising application of the Luminesensor is 3D printing, using light signals of spatially high resolution. Three dimensional images could be elegantly printed into media mixed with bacteria expressing the Luminesensor (see 3D Printing). A new design strategy of 3D printer using holographic technology instead of layer-by-layer printing, was also proposed. With such a power tool, 3D printing regarding biological material has endless potentials for medical use (crazy ideas in Future).
3. Cell-Cell Light Communication
-- A Novel Strategy of Signaling
Cell-cell communication based on quorum sensing systems, e.g. AHL, has been largely utilized by synthetic biologists to construct gene circuits with complex functions, e.g. pattern formation and edge detection. However, the diffusion and saturation of quorum sensing chemicals limit the delivery of communication signals, which in turn requires high spatiotemporal resolution and long range interaction.
The ultrasensitivity of the Luminesensor encouraged our team to explore the possibility of cell-cell light communication (Figure 4). By employing bacteria expressing the lux operon from V. fisheri as "light senders" and those bearing the Luminesensor as "light receiver", we successfully demonstrated, for the first time ever, that light communication between cells could be achieved beyond direct physical contact (see video in Results). Through measuring the dynamics of light communication, it was proved that bioluminescence was sufficient to trigger the response of the Luminesensor (data in Results).
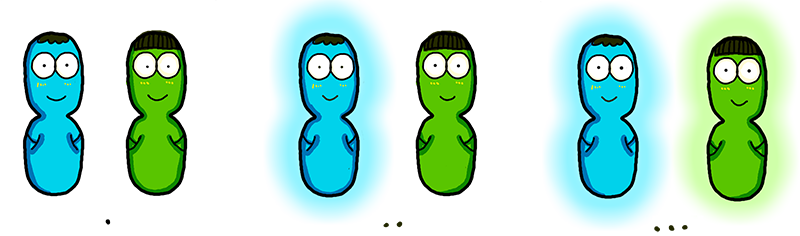
Figure 4.
Our Light-Off system could be upgraded into Light-On system through simple design (details in Design), which enables us to achieve more flexible cell-cell light communication (creative ideas in Future).
4. Phototaxis
--Controlling Cell Motion by Light
What makes Luminesensor outstanding is not only its ultra-sensitivity, but also the particularly high dynamic range (data on Characterization). Such switch-like property makes Luminesensor a very reliable and beneficial module in synthetic biology. Similar to the light-controlled animal behavior in neuroscience, by coupling Luminesensor with endogenous systems, it is possible to integrate the light signal with cell motion.
Figure 5.
"Phototatic" bacteria can be built by reprogramming the chemotaxis system in E. coli through light with the Luminesensor (Figure 5). By controlling the expression of the CheZ protein with light, the tumbling frequency is coupled to intensity of light signals (details in Design). On the border of the light and dark fields, motility ability of the cells in a single colony on the two sides is sufficient to result in an uneven colony (see Demonstration). Light-controlled cellular motion indeed has very promising environmental and medical applications for the future, e.g. the delivery of drugs to target concerns.
 "
"














