Team:Peking/Project/Luminesensor
From 2012.igem.org
Spring zhq (Talk | contribs) |
|||
| (30 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<div class="PKU_context floatR first"> | <div class="PKU_context floatR first"> | ||
<h3 class="title1">Introduction</h3> | <h3 class="title1">Introduction</h3> | ||
| - | <p>To date, equipped with optogenetic tools, scientists have been able to manipulate cellular behavior to achieve some truly fascinating goals, such as tuning the expression of genes through the intensity of light, making bacteria capable of detecting the edge of a light illuminated pattern, illuminating specific positions on the cell to trigger pseudopod formation or cell polarization, or even recruiting fluorescent proteins to cell membrane to print patterns on the cell.<br/><br/> | + | <p>To date, equipped with optogenetic tools, scientists have been able to manipulate cellular behavior to achieve some truly fascinating goals, such as tuning the expression of genes through the intensity of light, making bacteria capable of detecting the edge of a light illuminated pattern, illuminating specific positions on the cell to trigger pseudopod formation or cell polarization, or even recruiting fluorescent proteins to cell membrane to print patterns on the cell. |
| - | + | <br /><br /> | |
| - | However, as | + | However, the many advantages light possesses as a signal that can encode massive information have not yet been fully explored. As has illustrated in our project’s overview, the programmability, Bool-logic-like dynamic behavior and propagation that does not require any medium strongly propose that light may serve as an ideal signal for cell-cell communication that does not require physical contact. Such cell-cell communication has never been implemented artificially before in synthetic biology field. |
| - | + | <br /><br /> | |
| - | + | To realize this new-generation of cell-cell communication that must utilize the dim light emitted by cell itself as the carrier of information, we have to develop an ultra-sensitive sensor that possesses high on-off ratio and independency of exogenous chromophore both at the same time. | |
| - | + | <br /><br /> | |
| - | + | Under the invaluable instruction of <b>Prof. Yi Yang</b>, our ECUST team members designed a brand new prokaryotic light sensor following a general principle in optogenetic sensor design: to design a fusion protein with a photosensor domain preceding a physiological domain. Its photosensor domain is a VVD protein, the smallest photosensor protein known, which forms a rapidly exchanging dimer when excited by blue light and has been previously demonstrated to be very sensitive. Its physiologically functional domain is the DNA binding domain of the LexA protein, a bacteria transcription inhibitor, whose DNA binding activity rigorously requires the help of the dimerization domain. The VVD domain of this novel light sensor will form a dimer when absorbs blue light and help the DNA binding domain of LexA to dimerize and thus facilitate its DNA binding activity. So this novel light sensor will function as a blue light activated transcription repressor that will inhibit the expression of LexA responsive genes when exposed to blue light, generating a light-off system. (See <a href="https://2012.igem.org/Team:Peking/Project/Luminesensor/Design">Design</a>) | |
| - | + | <br /><br /></p> | |
| - | + | <img src="/wiki/images/1/1f/Peking2012_Design_illustration_of_function_mechanism_of_our_luminesensor.png" alt="Figure 9." style="width:600px;"/> | |
| + | <p class="description">Figure 1. Illustration of the function mechanism of the light-off system. When exposed to blue light, the N-terminal cap of VVD domain in the photosensor will undock and cause the VVD domain to dimerize. The dimerization of the VVD protein will help the dimerization of N-terminal domains of LexA protien fused to the N-terminal of the VVD domain. When helped to dimerize, the NTD of the LexA protein will recognize and bind to the SOS box in the promoter of our reporter gene and inhibit transcription initiation.</p> | ||
| + | <p>Having constructed the light-off system, we moved on to characterizing its sensitivity and on-off ratio. To our astonishment and satisfaction, we found that the brand new sensor was able to sense light as dim as bio-luminescence and respond in a switch-like manor with a dynamic range of over 400 fold. And we found it to be able to work properly in <i>E.coli</i> without any supply of exogenous chromophores. This light sensor was highly promising, for it was able to achieve high sensitivity, high on-off ratio and independence of exogenous chromophore all at the same time, fully fulfilling our original criteria. (See <a href="https://2012.igem.org/Team:Peking/Project/Luminesensor/Characterization">Characterization</a>). | ||
| + | <br /><br /> | ||
| + | However, the original light-off system is not orthogonal to the endogenous bacteria SOS system, sense any reporter gene of the light-off system will be inevitably inhibited by endogenous LexA protein. so the original light-off system will only be able to function in ΔLexA bacteria strains, which will greatly impede the light-off system’s future application. Fortunately, a LexA408 variant recognizes a different target sequence than the wild type LexA. Thus by reshuffling the LexA DNA binding domain to a 408 form, we would be able to ensure our light-off system’s orthogonality to the bacteria endogenous SOS system. (See <a href="https://2012.igem.org/Team:Peking/Project/Luminesensor/Characterization">Characterization</a>) | ||
| + | <br /><br /> | ||
| + | We use two LexA responsive promoters psulA and precA, and attached GFP reporter downstream of the 408 mutant form of these two promoters. We then tested them in both wild-type and ΔLexA <i>E.coli </i>strains. It proved that the mutated light-off system is perfectly orthogonal to the endogenous SOS system, while the wild type light-off system is severely interfered by the endogenous LexA protein. (See <a href="https://2012.igem.org/Team:Peking/Project/Luminesensor/Characterization">Characterization</a>)</p> | ||
| + | <div class="floatC"> | ||
| + | <img src="/wiki/images/e/e7/Peking2012_orthogonality_plate_group3.png" alt="Figure 6(3)." style="width:500px;"/> | ||
| + | <p class="description">Figure 2. Plate streaked with wild type <i>E.coli</i> transformed with mutated light-off system with GFP as a reporter gene under the control of precA408 and psulA408 promoter and cultivated in the dark or under light.(A: luminesensor + precA408-GFP, cultivated in the dark. B: luminesensor + precA408-GFP, cultivated under light. C: luminesensor + psulA408-GFP, cultivated in the dark. D: luminesensor + psulA408-GFP, cultivated under light.) Just as we predicted, the GFP reporter gene is expressed in the dark and supressed by our luminesensor under light. This clearly proved that mutated light-off system works orthogonally to host genetic context while retaining light-controllability.</p> | ||
| + | </div> | ||
| + | <p>Though the light-off system outshines many previously designed optogenetic modules, there is still space for improvements, such as the new light sensor’s slow decaying half-life.So we constructed a in-silico reaction network model for the light-off system’s light-induced transcription repression process, and conducted sensitive parameter analysis. </p> | ||
</div> | </div> | ||
</html> | </html> | ||
{{Template:Peking2012_Color_Epilogue}} | {{Template:Peking2012_Color_Epilogue}} | ||
Latest revision as of 13:54, 26 October 2012
Introduction
To date, equipped with optogenetic tools, scientists have been able to manipulate cellular behavior to achieve some truly fascinating goals, such as tuning the expression of genes through the intensity of light, making bacteria capable of detecting the edge of a light illuminated pattern, illuminating specific positions on the cell to trigger pseudopod formation or cell polarization, or even recruiting fluorescent proteins to cell membrane to print patterns on the cell.
However, the many advantages light possesses as a signal that can encode massive information have not yet been fully explored. As has illustrated in our project’s overview, the programmability, Bool-logic-like dynamic behavior and propagation that does not require any medium strongly propose that light may serve as an ideal signal for cell-cell communication that does not require physical contact. Such cell-cell communication has never been implemented artificially before in synthetic biology field.
To realize this new-generation of cell-cell communication that must utilize the dim light emitted by cell itself as the carrier of information, we have to develop an ultra-sensitive sensor that possesses high on-off ratio and independency of exogenous chromophore both at the same time.
Under the invaluable instruction of Prof. Yi Yang, our ECUST team members designed a brand new prokaryotic light sensor following a general principle in optogenetic sensor design: to design a fusion protein with a photosensor domain preceding a physiological domain. Its photosensor domain is a VVD protein, the smallest photosensor protein known, which forms a rapidly exchanging dimer when excited by blue light and has been previously demonstrated to be very sensitive. Its physiologically functional domain is the DNA binding domain of the LexA protein, a bacteria transcription inhibitor, whose DNA binding activity rigorously requires the help of the dimerization domain. The VVD domain of this novel light sensor will form a dimer when absorbs blue light and help the DNA binding domain of LexA to dimerize and thus facilitate its DNA binding activity. So this novel light sensor will function as a blue light activated transcription repressor that will inhibit the expression of LexA responsive genes when exposed to blue light, generating a light-off system. (See Design)
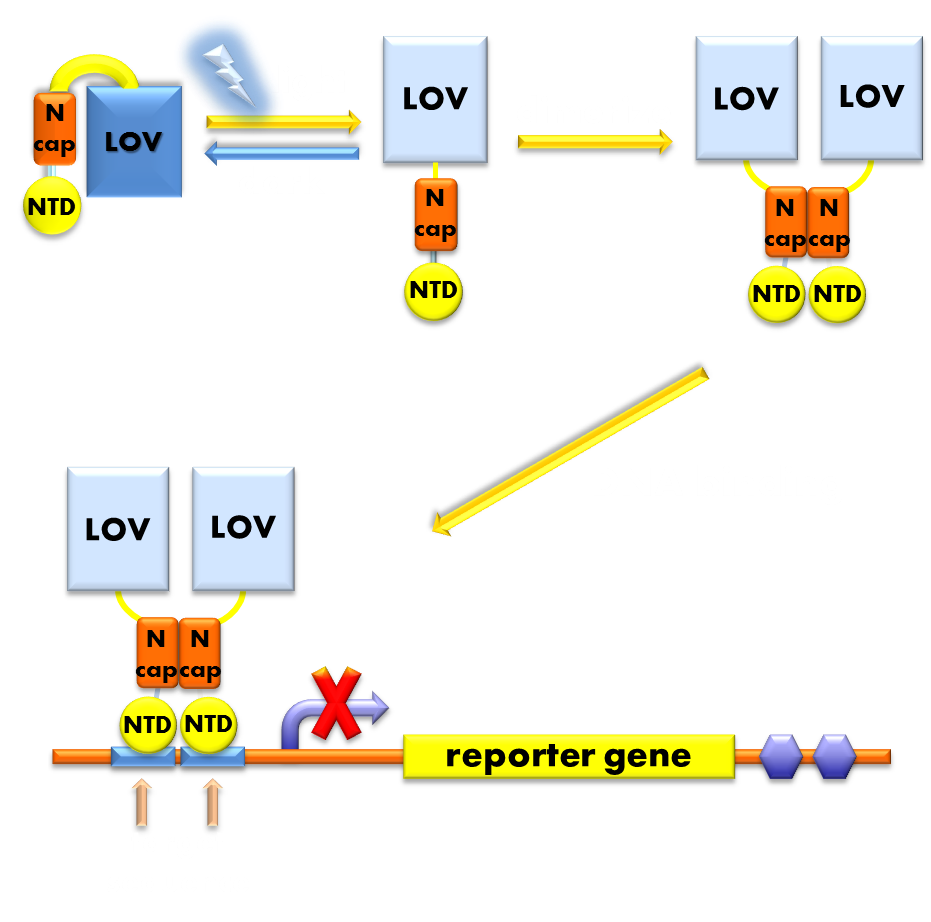
Figure 1. Illustration of the function mechanism of the light-off system. When exposed to blue light, the N-terminal cap of VVD domain in the photosensor will undock and cause the VVD domain to dimerize. The dimerization of the VVD protein will help the dimerization of N-terminal domains of LexA protien fused to the N-terminal of the VVD domain. When helped to dimerize, the NTD of the LexA protein will recognize and bind to the SOS box in the promoter of our reporter gene and inhibit transcription initiation.
Having constructed the light-off system, we moved on to characterizing its sensitivity and on-off ratio. To our astonishment and satisfaction, we found that the brand new sensor was able to sense light as dim as bio-luminescence and respond in a switch-like manor with a dynamic range of over 400 fold. And we found it to be able to work properly in E.coli without any supply of exogenous chromophores. This light sensor was highly promising, for it was able to achieve high sensitivity, high on-off ratio and independence of exogenous chromophore all at the same time, fully fulfilling our original criteria. (See Characterization).
However, the original light-off system is not orthogonal to the endogenous bacteria SOS system, sense any reporter gene of the light-off system will be inevitably inhibited by endogenous LexA protein. so the original light-off system will only be able to function in ΔLexA bacteria strains, which will greatly impede the light-off system’s future application. Fortunately, a LexA408 variant recognizes a different target sequence than the wild type LexA. Thus by reshuffling the LexA DNA binding domain to a 408 form, we would be able to ensure our light-off system’s orthogonality to the bacteria endogenous SOS system. (See Characterization)
We use two LexA responsive promoters psulA and precA, and attached GFP reporter downstream of the 408 mutant form of these two promoters. We then tested them in both wild-type and ΔLexA E.coli strains. It proved that the mutated light-off system is perfectly orthogonal to the endogenous SOS system, while the wild type light-off system is severely interfered by the endogenous LexA protein. (See Characterization)
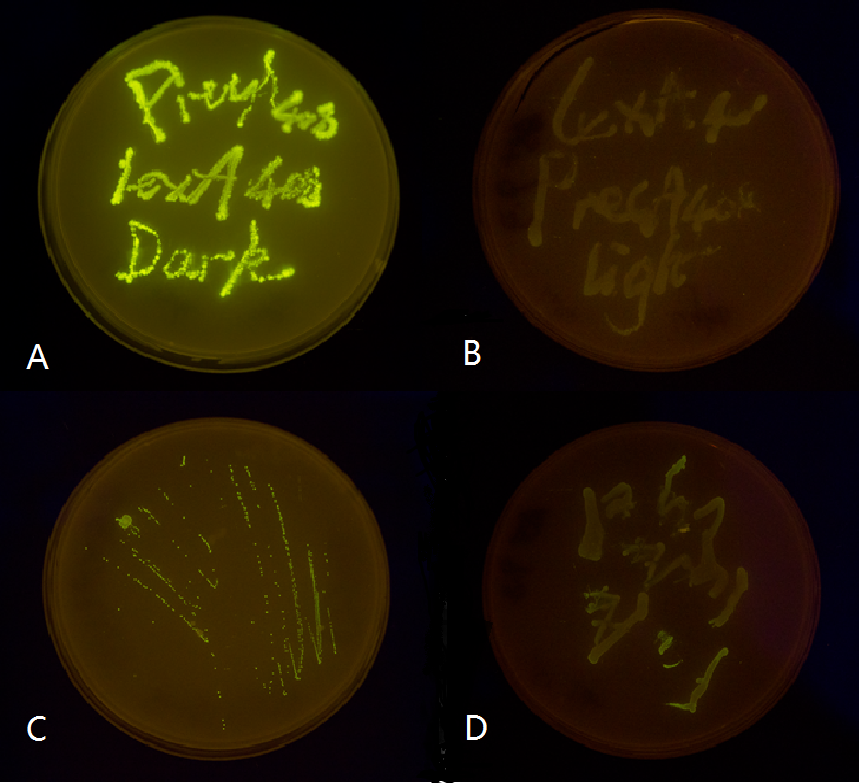
Figure 2. Plate streaked with wild type E.coli transformed with mutated light-off system with GFP as a reporter gene under the control of precA408 and psulA408 promoter and cultivated in the dark or under light.(A: luminesensor + precA408-GFP, cultivated in the dark. B: luminesensor + precA408-GFP, cultivated under light. C: luminesensor + psulA408-GFP, cultivated in the dark. D: luminesensor + psulA408-GFP, cultivated under light.) Just as we predicted, the GFP reporter gene is expressed in the dark and supressed by our luminesensor under light. This clearly proved that mutated light-off system works orthogonally to host genetic context while retaining light-controllability.
Though the light-off system outshines many previously designed optogenetic modules, there is still space for improvements, such as the new light sensor’s slow decaying half-life.So we constructed a in-silico reaction network model for the light-off system’s light-induced transcription repression process, and conducted sensitive parameter analysis.
 "
"














