Team:Peking/Project
From 2012.igem.org
Synthetic Biology: What's Hindering Us?
Efforts in synthetic biology have been primarily focused on the creation and perfection of biological modules and systems in order to achieve specific functions. Since first demonstrated by the bacterial toggle switch and the Repressilator in 2000, synthetic biologists now possess the ability to engineer and manipulate genetic circuits, metabolic pathways, and even entire genomes.
In genetic engineering, chemicals are frequently employed as signals to control molecular and cellular behavior. However, because chemicals deliver information only by diffusion, chemical signals are seriously limited by the short-range interactions. Another serious issue regarding chemical signals is the complexity of the process in fully eradicating them, making it difficult for the system to reset. Additionally, the cytotoxicity, high cost, and narrow dynamic range of chemicals urge synthetic biologists to find more spatiotemporally specific and environmental friendly alternatives.
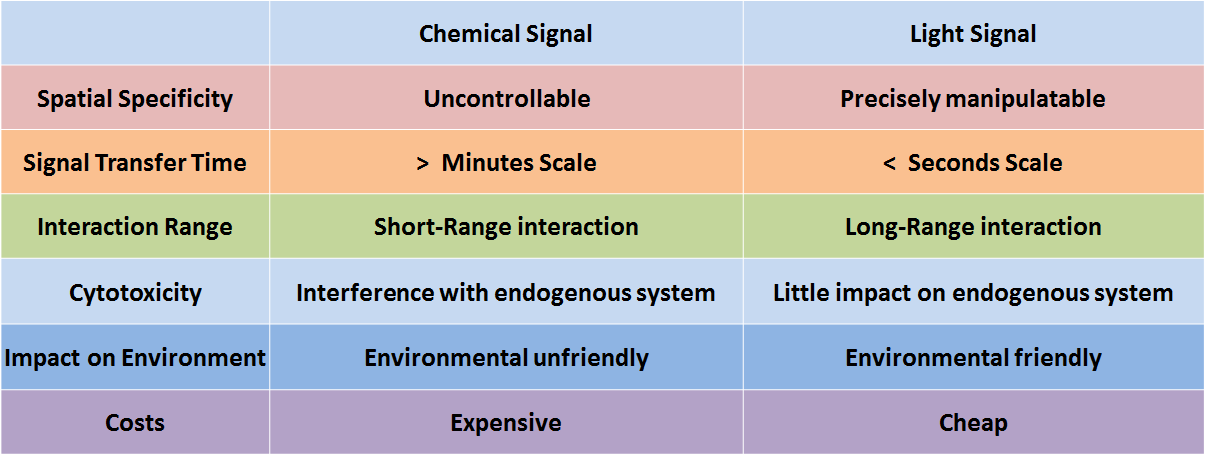
Figure 1. Comparison of chemical signal and light signal
Optogenetics: Inspire Synthetic Biology
Literally, optogenetics refers to the genetic strategies which employ light as the input signal in the place of chemical signals. During the last decade, optogenetics has made significant impact on life sciences and other practices. It has evolved rapidly, with light-switchable promoters, light-induced protein-protein interaction, light-activated ion channels and eventually, light-controlled animal behavior, and so on. The spatiotemporal specificity of light signals allows for precise manipulation and long range interaction without physical contact. Besides its incredible range of functions, light resources are also cheaper, more sustainable, and environmentally friendly. These properties together make optogenetic tools attractive for synthetic biologists.
![[Fig 2.]](/wiki/images/a/aa/Peking2012_Optogenetics.jpg)
Figure 2. Evolution of Optogenetics.
However, current optogenetic tools are far from satisfactory for synthetic biology applications. In the next section, we will systematically analyze the current optogenetic methods and raise issues that our Luminesensor needs to circumvent.
Issues of Current Optogenetic Methods
We carefully selected 15 representative optogenetic methods, ranging from those used in prokaryotes to those in eukaryotes. By statistically evaluating the modularity, sensitivity, and dynamic range of these optogenetics methods, we have come to address these four issues of current optogenetics approaches:
![[Fig 3.]](/wiki/images/5/5b/Peking2012_effect_of_laser_1.jpg)
Figure 3a. The inhibitory effect of lasers on bacterial growth.
1. Low Sensitivity
To minimize the influence on cell growth, strong light needs to be avoided for practical applications. Nearly 50% of current 15 optogenetic tools employ lasers as the light source. We demonstrated the lethal effect of lasers by illuminating a lawn of E.coli with a laser pointer. As clearly shown in Figure 2, no bacterial growth could be found on the spot that was illuminated. Only about 25% of the 15 optogenetics tools are able to sense natural light (~1W/m2), a biologically friendly and much more sustainable source of light (Figure 3a, b).
2. Narrow Dynamic Range
For a control system, the dynamic range (fold change of output in response to input signal) is an essential parameter used to evaluate the control efficiency. Among the 15 optogenetic methods, few have a dynamic range that reaches 100-fold (Figure 3b). In application, systems with narrow dynamic ranges will be easily interfered by the intrinsic and extrinsic noises. The main motive for having narrow dynamic range is due to the fact that most of the methods utilize a complex signal cascade where high basal level is inevitable.
![[Fig 3b.]](/wiki/images/e/ee/Peking2012_YL_overview_sensitivity_dynamic_range.png)
Figure 3b. The sensitivity and dynamic range of the 15 optogenetic methods.
3. Dependency on Exogenous Chromophores
Many optogenetic methods require the supplement of exogenous chromophores, e.g. phycocyanobilin (PCB) and caged amino acids. This leads to two major problems:
(1) The exogenous chromophores may be incompatible with the endogenous components, and observed in some cases, even have cytotoxity;
(2) The uneven diffusion of chromophores, in many cases, makes the output unpredictable.
4. Limited Application in Prokaryotes
Another serious issue concerns the limited application of optogenetic methods in prokaryotes. The rapidly reproducing prokaryote plays a very significant role in both fundamental research and industrial application of synthetic biology. In many methods, only proof of concept is provided without considering practical application.
Low sensitivity, narrow dynamic range, and the need for exogenous chromophores, as mentioned above, are the obstacles for a broader and more practical application of optogenetics in synthetic biology.
A New Generation of Optogenetic Tool:
The Luminesensor
By gaining insight into the current optogenetic methods, Peking iGEM team speculates that rationally designing a robust and ultra-sensitive transcription regulator that works in prokaryotes will circumvent all the issues mentioned above. High sensitivity allows for dimmer, cheaper, and safer light as the source of signals. Because the light sensitive transcription regulator functions directly on the gene expression, the basal level will also be low enough to guarantee a high dynamic range.
Indeed, as you will see later, such an ultrasensitive and efficient optogenetic tool has been successfully designed. Moreover, we endeavored to explore the full proficiency of our Luminesensor and raised a new generation of optogenetic tool with broader applications on the manipulation of biochemical process, cellular behavior, and even, inter-cellular communication.
Reference
-
1. Wu, Y. I., D. Frey, et al. (2009). "A genetically encoded photoactivatable Rac controls the motility of living cells." Nature 461(7260): 104-108.
2.Shimizu-Sato, S., E. Huq, et al. (2002). "A light-switchable gene promoter system." Nat Biotechnol 20(10): 1041-1044.
3.Moglich, A., R. A. Ayers, et al. (2009). "Design and signaling mechanism of light-regulated histidine kinases." J Mol Biol
4.Gradinaru, V., F. Zhang, et al. (2010). "Molecular and cellular approaches for diversifying and extending optogenetics." Cell 141(1): 154-165.
5.Ohlendorf, R., R. R. Vidavski, et al. (2012). "From dusk till dawn: one-plasmid systems for light-regulated gene expression." J Mol Biol 416(4): 534-542.
6.Miesenbock, G. (2011). "Optogenetic control of cells and circuits." Annu Rev Cell Dev Biol 27: 731-758.
7.Zemelman, B. V., G. A. Lee, et al. (2002). "Selective photostimulation of genetically chARGed neurons." Neuron 33(1): 15-22.
8.Levskaya, A., O. D. Weiner, et al. (2009). "Spatiotemporal control of cell signalling using a light-switchable protein interaction." Nature 461(7266): 997-1001.
9.Toettcher, J. E., C. A. Voigt, et al. (2011). "The promise of optogenetics in cell biology: interrogating molecular circuits in space and time." Nat Methods 8(1): 35-38.
 "
"














