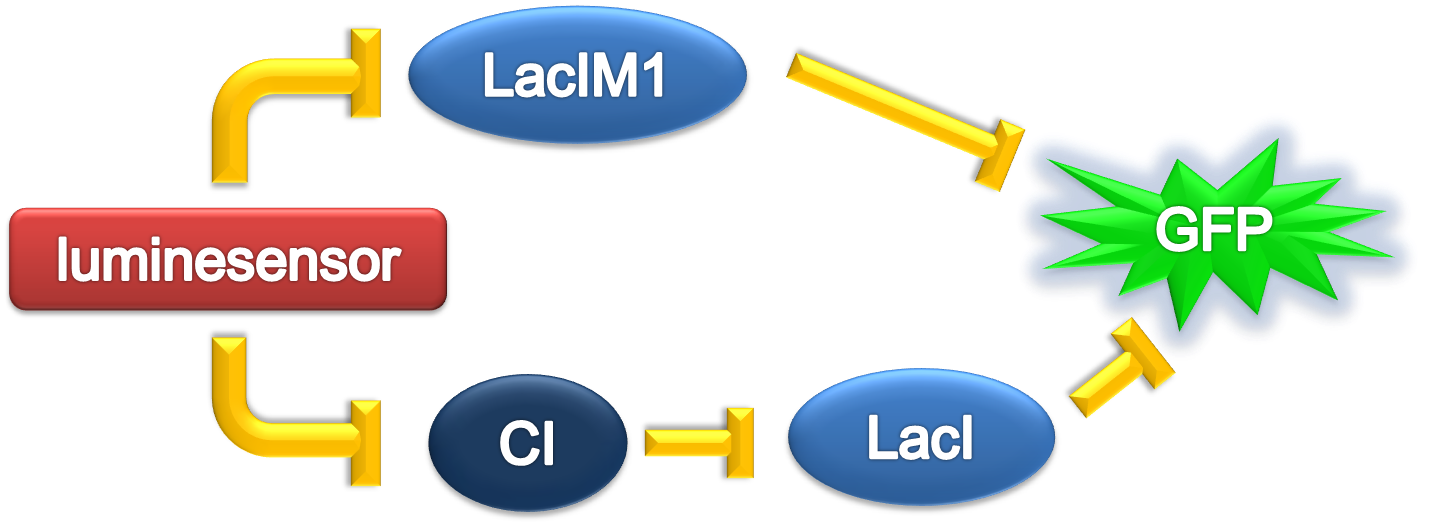
Team:Peking/Modeling/Ring/Simulation
From 2012.igem.org
| Line 27: | Line 27: | ||
<td>Parameter</td><td>Value</td><td>Unit</td><td>Description</td><td>Source</td> | <td>Parameter</td><td>Value</td><td>Unit</td><td>Description</td><td>Source</td> | ||
</tr><tr> | </tr><tr> | ||
| - | <td>a<sub>G</sub></td><td>2</td><td>10<sup>-6</sup>M/min</td><td> | + | <td>a<sub>G</sub></td><td>2</td><td>10<sup>-6</sup>M/min</td><td>GFP production rate constant</td><td></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>a<sub>C</sub></td><td>2</td><td>10<sup>-6</sup>M/min</td><td> | + | <td>a<sub>C</sub></td><td>2</td><td>10<sup>-6</sup>M/min</td><td>CI production rate constant</td><td><a href="#ref3" title="Zoltowski, B.D., Vaccaro, B., and Crane, B.R. (2009). Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol. 5: 827: 834">[3]</a></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>a<sub>L1</sub></td><td>1</td><td>10<sup>-6</sup>M/min</td><td> | + | <td>a<sub>L1</sub></td><td>1</td><td>10<sup>-6</sup>M/min</td><td>LacI production rate constant</td><td></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>a<sub>L2</sub></td><td>1</td><td>10<sup>-6</sup>M/min</td><td> | + | <td>a<sub>L2</sub></td><td>1</td><td>10<sup>-6</sup>M/min</td><td>LacIM1 production rate constant</td><td></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>b<sub>C</sub></td><td>8.x10<sup>-3</sup></td><td>10<sup>-6</sup>M</td><td> | + | <td>b<sub>C</sub></td><td>8.x10<sup>-3</sup></td><td>10<sup>-6</sup>M</td><td>Binding strength of CI on LacI operator</td><td></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>b<sub>L</sub></td><td>8.x10<sup>-1</sup></td><td>10<sup>-6</sup>M</td><td> | + | <td>b<sub>L</sub></td><td>8.x10<sup>-1</sup></td><td>10<sup>-6</sup>M</td><td>Binding strength of LacI or LacIM1 on GFP operator</td><td></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>b<sub>R</sub></td><td>1.x10<sup>-2</sup></td><td>10<sup>-6</sup>M</td><td> | + | <td>b<sub>R</sub></td><td>1.x10<sup>-2</sup></td><td>10<sup>-6</sup>M</td><td>Binding strength of <i>Luminesensor</i> on corresponding operator</td><td></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>r<sub>G</sub></td><td>6.92x10<sup>-2</sup></td><td>min<sup>-1</sup></td><td> | + | <td>r<sub>G</sub></td><td>6.92x10<sup>-2</sup></td><td>min<sup>-1</sup></td><td>GFP dissociation rate constant</td><td><a href="#ref1" title="Zoltowski, B.D., Crane, B.R.(2008). Light Activation of the LOV Protein Vivid Generates a Rapidly Exchanging Dimer.Biochemistry, 47: 7012: 7019 ">[1]</a></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>r<sub>C</sub></td><td>6.92x10<sup>-2</sup></td><td>min<sup>-1</sup></td><td> | + | <td>r<sub>C</sub></td><td>6.92x10<sup>-2</sup></td><td>min<sup>-1</sup></td><td>CI dissociation rate constant</td><td><a href="#ref2" title="2. Mohana-Borges, R., Pacheco, A.B., Sousa, F.J., Foguel, D., Almeida, D.F., and Silva, J.L. (2000). LexA repressor forms stable dimers in solution. The role of specific DNA in tightening protein-protein interactions. J. Biol. Chem., 275: 4708: 4712">[2]</a></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>r<sub>L</sub></td><td>2.31x10<sup>-2</sup></td><td>min<sup>-1</sup></td><td> | + | <td>r<sub>L</sub></td><td>2.31x10<sup>-2</sup></td><td>min<sup>-1</sup></td><td>LacI and LacIM1 dissociation rate constant</td><td>Thermal Principle</td> |
</tr><tr> | </tr><tr> | ||
| - | <td>r<sub>R</sub></td><td>2.31x10<sup>-2</sup></td><td>min<sup>-1</sup></td><td> | + | <td>r<sub>R</sub></td><td>2.31x10<sup>-2</sup></td><td>min<sup>-1</sup></td><td><i>Luminesensor</i> dissociation rate constant</td><td><a href="#ref2" title="Mohana-Borges, R., Pacheco, A.B., Sousa, F.J., Foguel, D., Almeida, D.F., and Silva, J.L. (2000). LexA repressor forms stable dimers in solution. The role of specific DNA in tightening protein-protein interactions. J. Biol. Chem., 275: 4708: 4712">[2]</a></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>I<sub>0</sub></td><td>1000</td><td>AU</td><td> | + | <td>I<sub>0</sub></td><td>1000</td><td>AU</td><td>Maximum light intensity in the middle of the plate </td><td></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>k</td><td>500</td><td>10< | + | <td>k</td><td>500</td><td>10<sup>-6</sup>M</td><td><i>Luminesensor</i> activation rate under light</td><td></td> |
</tr><tr> | </tr><tr> | ||
| - | <td>K</td><td>10000</td><td>AU</td><td> | + | <td>K</td><td>10000</td><td>AU</td><td>light sensitivity of <i>Luminesensor</i> activation </td><td></td> |
</tr> | </tr> | ||
</table> | </table> | ||
Revision as of 04:07, 24 October 2012
ODE Model
According to the previous circuit and ODE model, we listed all the differential equations and simulated this system with MATLAB with equations listed as below:
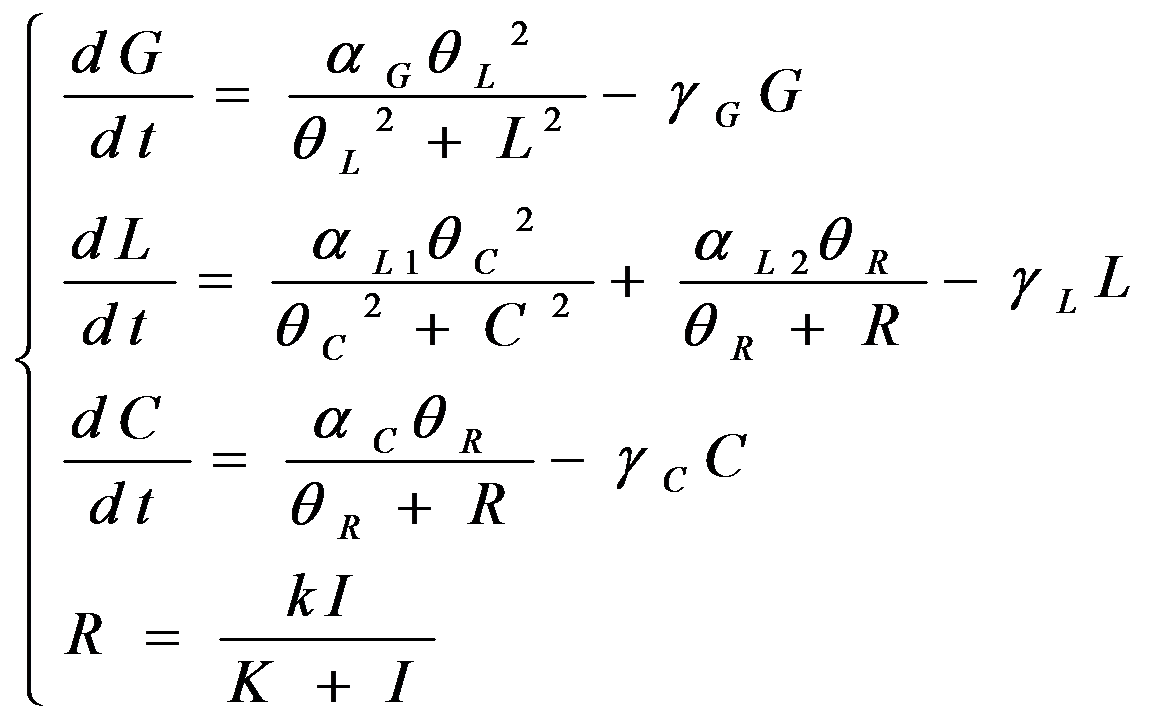
And parameters as
| Parameter | Value | Unit | Description | Source |
| aG | 2 | 10-6M/min | GFP production rate constant | |
| aC | 2 | 10-6M/min | CI production rate constant | [3] |
| aL1 | 1 | 10-6M/min | LacI production rate constant | |
| aL2 | 1 | 10-6M/min | LacIM1 production rate constant | |
| bC | 8.x10-3 | 10-6M | Binding strength of CI on LacI operator | |
| bL | 8.x10-1 | 10-6M | Binding strength of LacI or LacIM1 on GFP operator | |
| bR | 1.x10-2 | 10-6M | Binding strength of Luminesensor on corresponding operator | |
| rG | 6.92x10-2 | min-1 | GFP dissociation rate constant | [1] |
| rC | 6.92x10-2 | min-1 | CI dissociation rate constant | [2] |
| rL | 2.31x10-2 | min-1 | LacI and LacIM1 dissociation rate constant | Thermal Principle |
| rR | 2.31x10-2 | min-1 | Luminesensor dissociation rate constant | [2] |
| I0 | 1000 | AU | Maximum light intensity in the middle of the plate | |
| k | 500 | 10-6M | Luminesensor activation rate under light | |
| K | 10000 | AU | light sensitivity of Luminesensor activation |
The simulation result is shown below:
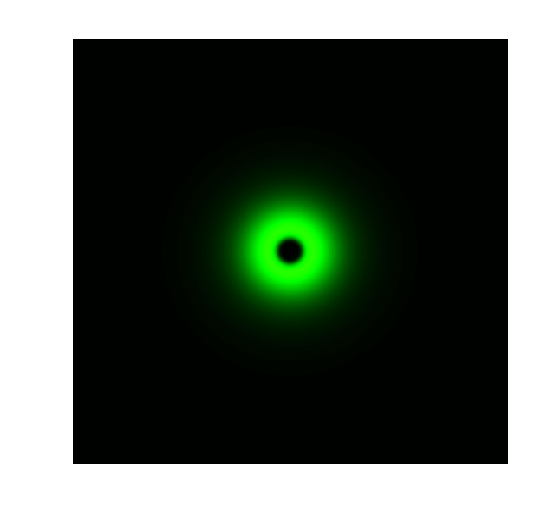
Figure 1. ODE Simulation in a plate of the ring-like pattern formation.
Figure 2. ODE Simulation for the radial expression amplitude of the ring-like pattern formation.
From the Figure 1 above, we discovered that the activation and decay of Luminesensor are the key points of progress, and the activating rate is the most sensitive to light intensity. The promoter will be repressed even though the Luminesensor does not totally dimerized.
Parameter Analysis
After modeling the prototype Luminesensor, we attempted to optimize it in a rational way. We have tuned the parameters both up and down, one by one, and finally discovered four parameters which predominantly influence the performance of the Luminesensor.
| Function | Parameter | Description | Remark |
| Reduce responsing time | k1 | Vivid lighting decay rate constant | Mainly on process from Light to Dark |
| k3 | rate constant of monomer LexA releasing from specific binding site | ||
| Enhance contrast | K2 | Vivid association equilibrium constant | More dimerization provides more binding opportunity |
| K5 | dimered LexA binding equilibrium constant | More binding affinity |
Reference
- 1. Subhayu Basu et al.(2005), A synthetic multicellular system for programmed pattern formation. Nature, vol.434: 1130: 1134
 "
"














