Team:Peking
From 2012.igem.org
m |
|||
| Line 869: | Line 869: | ||
<div class="floatR"><img src="/wiki/images/a/a8/Peking2012_PhototaxisSPECSdot_Edit.gif" alt="" style="width:200px;"/></div> | <div class="floatR"><img src="/wiki/images/a/a8/Peking2012_PhototaxisSPECSdot_Edit.gif" alt="" style="width:200px;"/></div> | ||
<div class="floatC"><p class="context_block" style="width:360px;"> | <div class="floatC"><p class="context_block" style="width:360px;"> | ||
| - | + | Excellent stochastic simulation was conducted to rationally optimize our <i>Luminesensor</i>, combining protein kinetics and reaction thermodynamics. Molecular docking was also operated. What's more, while designing our photo-taxis model, we developed a hexagonal-coordinate environment for dynamic simulation of “on-plate" system. Those who focus on cell motility will find it useful and get inspired. | |
</p></div> | </p></div> | ||
</div> | </div> | ||
Revision as of 16:05, 26 September 2012
O
ptogenetic tools have made significant impact on life sciences and beyond. However, several serious issues remain: cytotoxicity, narrow dynamic range, and dependency on laser and exogenous chromophores. To unblock the bottleneck, Peking iGEM has rationally constructed a hypersensitive sensor of luminance -- Luminesensor. Primarily, the sensor was designed by fusing the blue-light-sensing protein domain from Neurospora with the DNA binding domain of LexA from E. coli. Subsequently, protein structure inspection and kinetic simulation were conducted to rationally perform optimization. Amazingly, Luminesensor was proved to be so sensitive as to sense natural light and even bioluminescence. With this sensor, spatiotemporal control of cellular behavior, such as high-resolution 2D and 3D bio-printing using dim light and even luminescence of iPad were shown to be very easy. What’s more, we successfully implemented cell-cell signaling using light, which is the very first time in synthetic biology and of great importance for biotechnological use.
Abstract |
Peking iGEM would never disappoint you!
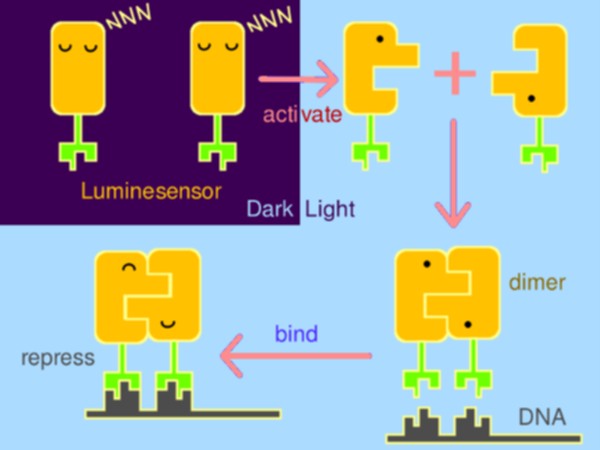
Strongly motivated to open up a new era of optogenetics, Peking iGEM team has rationally constructed a hypersensitive sensor of luminance – named by us Luminesensor, enabling highly efficient spatiotemporal control of biochemical process or cellular behavior. The whole job serves as a paradigm for sensor creating.

The hypersensitive Luminesensor is able to respond to very low light and meanwhile keep a wide dynamic range, which encouraged Peking iGEM to explore the possibility of cell-cell communication through light. We have successfully implemented, for the very first time, light-communication among cells without direct physical contact.

3D printing is a new technology that has been rising for many years. But in the realm of synthetic biology, it is far from developed. We exploited our Luminesensor to implement 3D printing that can be utilized in many applications in medical or manufacturing.
"Phototatic" bacteria can be built by programming the chemotaxis system in E. coli through light. By controlling the expression level of the CheZ protein with Luminesensor, the tumbling frequency is coupled to the intensity of light signals.

We have done a remarkable job in motivating high school students to study synthetic biology and guiding them towards future participation in the iGEM high school division. Besides, we collaborated with a lab and helped 4 other iGEM teams by sharing DNA materials, characterizing their parts and modeling. What's more, we presented all fresh iGEMers with a collection and praise of historic iGEM projects to share and learn from each other!
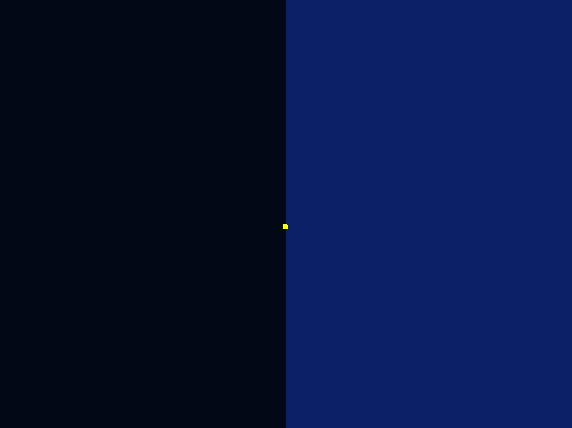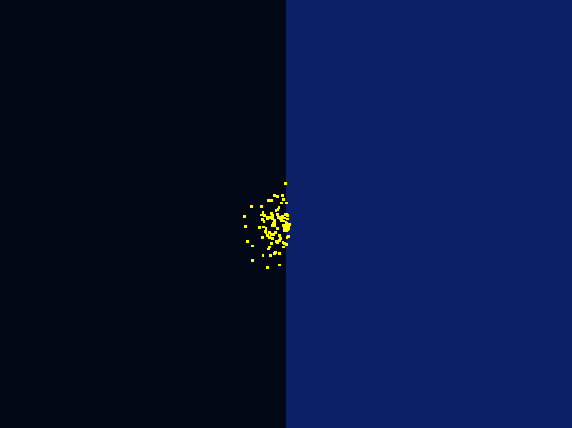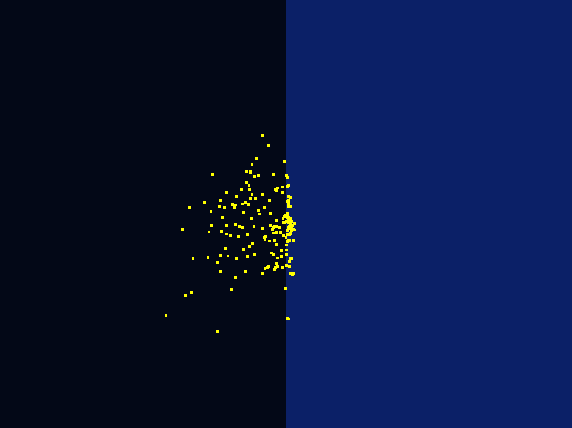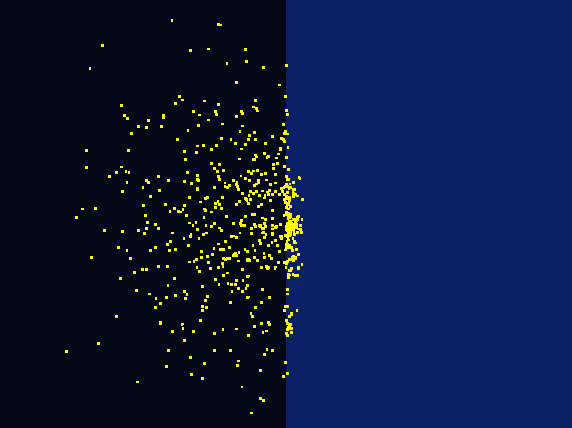
Excellent stochastic simulation was conducted to rationally optimize our Luminesensor, combining protein kinetics and reaction thermodynamics. Molecular docking was also operated. What's more, while designing our photo-taxis model, we developed a hexagonal-coordinate environment for dynamic simulation of “on-plate" system. Those who focus on cell motility will find it useful and get inspired.
 "
"















