Team:Peking
From 2012.igem.org
m |
|||
| Line 509: | Line 509: | ||
</li> | </li> | ||
<li style="width:75px;"> | <li style="width:75px;"> | ||
| - | <a onmouseover="TriggerSubmenu(1);" onmouseout="TriggerSubmenu(0); | + | <a onmouseover="TriggerSubmenu(1);" onmouseout="TriggerSubmenu(0);"> |
Team | Team | ||
</a> | </a> | ||
</li> | </li> | ||
<li style="width:85px;"> | <li style="width:85px;"> | ||
| - | <a onmouseover="TriggerSubmenu(2);" onmouseout="TriggerSubmenu(0); | + | <a onmouseover="TriggerSubmenu(2);" onmouseout="TriggerSubmenu(0);"> |
Project | Project | ||
</a> | </a> | ||
</li> | </li> | ||
<li style="width:100px;"> | <li style="width:100px;"> | ||
| - | <a onmouseover="TriggerSubmenu( | + | <a onmouseover="TriggerSubmenu(3);" onmouseout="TriggerSubmenu(0);"> |
Data Page | Data Page | ||
</a> | </a> | ||
</li> | </li> | ||
<li style="width:95px;"> | <li style="width:95px;"> | ||
| - | <a onmouseover="TriggerSubmenu( | + | <a onmouseover="TriggerSubmenu(4);" onmouseout="TriggerSubmenu(0);"> |
Modeling | Modeling | ||
</a> | </a> | ||
</li> | </li> | ||
<li style="width:145px;"> | <li style="width:145px;"> | ||
| - | <a onmouseover="TriggerSubmenu( | + | <a onmouseover="TriggerSubmenu(5);" onmouseout="TriggerSubmenu(0);"> |
Human Practice | Human Practice | ||
</a> | </a> | ||
| Line 547: | Line 547: | ||
<li> | <li> | ||
<ul onmouseover="TriggerSubmenu(1);" onmouseout="TriggerSubmenu(0);" class="vertical" style="width:120px;display:block;"> | <ul onmouseover="TriggerSubmenu(1);" onmouseout="TriggerSubmenu(0);" class="vertical" style="width:120px;display:block;"> | ||
| + | <li class="submenu_team"> | ||
| + | <a href="/Team:Peking/Team/History"> | ||
| + | Gallery | ||
| + | </a> | ||
| + | </li> | ||
<li class="submenu_team"> | <li class="submenu_team"> | ||
<a href="/Team:Peking/Team/Members"> | <a href="/Team:Peking/Team/Members"> | ||
| Line 560: | Line 565: | ||
<a href="/Team:Peking/Team/Diary"> | <a href="/Team:Peking/Team/Diary"> | ||
Diary | Diary | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</a> | </a> | ||
</li> | </li> | ||
| Line 571: | Line 571: | ||
<li> | <li> | ||
<ul onmouseover="TriggerSubmenu(2);" onmouseout="TriggerSubmenu(0);" class="vertical" style="width:200px;display:block;"> | <ul onmouseover="TriggerSubmenu(2);" onmouseout="TriggerSubmenu(0);" class="vertical" style="width:200px;display:block;"> | ||
| + | <li class="submenu_project"> | ||
| + | <a href="/Team:Peking/Project/Luminesensor"> | ||
| + | Overview | ||
| + | </a> | ||
| + | </li> | ||
<li class="submenu_project"> | <li class="submenu_project"> | ||
<a href="/Team:Peking/Project/Luminesensor"> | <a href="/Team:Peking/Project/Luminesensor"> | ||
| Line 594: | Line 599: | ||
</li> | </li> | ||
<li> | <li> | ||
| - | <ul onmouseover="TriggerSubmenu(3);" onmouseout="TriggerSubmenu(0);" class="vertical" style="width:250px;display:block;"> | + | <ul onmouseover="TriggerSubmenu(3);" onmouseout="TriggerSubmenu(0);" class="vertical" style="width:170px;display:block;"> |
| + | <li class="submenu_project"> | ||
| + | <a href="/Team:Peking/DataPage"> | ||
| + | BioBrick Data | ||
| + | </a> | ||
| + | </li> | ||
| + | <li class="submenu_project"> | ||
| + | <a href="/Team:Peking/DataPage/Judging"> | ||
| + | Judging Criteria | ||
| + | </a> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <ul onmouseover="TriggerSubmenu(4);" onmouseout="TriggerSubmenu(0);" class="vertical" style="width:250px;display:block;"> | ||
<li class="submenu_modeling"> | <li class="submenu_modeling"> | ||
<a href="/Team:Peking/Modeling/Luminesensor"> | <a href="/Team:Peking/Modeling/Luminesensor"> | ||
| Line 606: | Line 625: | ||
</li> | </li> | ||
<li class="submenu_modeling"> | <li class="submenu_modeling"> | ||
| - | <a href="/Team:Peking/Modeling/ | + | <a href="/Team:Peking/Modeling/Appendix/ODE"> |
Appendix | Appendix | ||
</a> | </a> | ||
| Line 625: | Line 644: | ||
</li> | </li> | ||
<li class="submenu_humanpractice"> | <li class="submenu_humanpractice"> | ||
| - | <a href="/Team:Peking/HumanPractice/Outreach"> | + | <a href="/Team:Peking/HumanPractice/Outreach/Visiting"> |
Outreach & Workshop | Outreach & Workshop | ||
</a> | </a> | ||
| Line 642: | Line 661: | ||
*/ | */ | ||
| - | var Submenu_Size_Cur = [ | + | var Submenu_Size_Cur = [130, 0, 0, 0, 0, 0]; |
| - | var Submenu_Item_Num = | + | var Submenu_Item_Num = 5; |
var Submenu_Item_Des = 0; | var Submenu_Item_Des = 0; | ||
var Submenu_Item_Cur = 0; | var Submenu_Item_Cur = 0; | ||
| - | var Submenu_BlankWidth = [0, 318, 360, 528, 612]; //Unit: Pixel | + | var Submenu_BlankWidth = [0, 318, 360, 400, 528, 612]; //Unit: Pixel |
| - | var Submenu_ListHeight = [ | + | var Submenu_ListHeight = [130, 100, 125, 50, 75, 75]; //Unit: Pixel |
var Submenu_Handle_State = 0; //0:Null, 1:Growth, 2:Waiting, 3:Wither | var Submenu_Handle_State = 0; //0:Null, 1:Growth, 2:Waiting, 3:Wither | ||
var Submenu_Handle = null; | var Submenu_Handle = null; | ||
| Line 789: | Line 808: | ||
</script> | </script> | ||
<div id="text-body"> | <div id="text-body"> | ||
| - | <div class=" | + | <div class="floatL" style="margin:25px 10px 5px 20px;"><table class="title_block" style="background-image:url('/wiki/images/b/be/Peking2012_Color_Title_AquaBlue.jpg');"><tr><td style="height:165px;"><p style="font-size:36px;">Abstract</p></td></tr></table></div> |
| - | + | <div class="floatL" style="margin:5px 10px 5px 20px;"><p style="font-size:36px;font-family:Verdana;"><b>HERE HERE</b></div> | |
| + | <div class="floatR"> | ||
<div style="float:left;"><p style="font-size:45px;line-height:45px;color:#ffffff;margin:20px 3px 0;">O</p></div> | <div style="float:left;"><p style="font-size:45px;line-height:45px;color:#ffffff;margin:20px 3px 0;">O</p></div> | ||
| - | <p class="context_block" style="width: | + | <p class="context_block" style="width:604px;"> |
ptogenetic tools have made significant impact on life sciences and beyond. However, several serious issues remain: cytotoxicy, narrow dynamic range, and dependency on laser and exogenous chromophore. To circumvent these, Peking iGEM has rationally constructed a hypersensitive sensor of luminance -- <b><i>Luminesensor</i></b>. Primarily, the sensor was designed by fusing blue-light-sensing protein domain from <i>Neurospora</i> with DNA binding domain of LexA from <i>E. coli</i>, following which protein structure inspection and kinetic simulation were conducted to rationally perform optimization. Amazingly, <i>Luminesensor</i> was proved to be as sensitive as to sense <b>natural light</b> and even <b>bioluminescence</b>. With this sensor, spatiotemporal control of cellular behavior, such as <b>high-resolution 2D</b> and <b>3D bio-printing</b> using dim light and even <b>luminescence of iPad</b> were shown to be very easy. What’s more, we successfully implemented <b>cell-cell signaling using light</b>, which is the very first time in synthetic biology and of great importance for biotechnological use. | ptogenetic tools have made significant impact on life sciences and beyond. However, several serious issues remain: cytotoxicy, narrow dynamic range, and dependency on laser and exogenous chromophore. To circumvent these, Peking iGEM has rationally constructed a hypersensitive sensor of luminance -- <b><i>Luminesensor</i></b>. Primarily, the sensor was designed by fusing blue-light-sensing protein domain from <i>Neurospora</i> with DNA binding domain of LexA from <i>E. coli</i>, following which protein structure inspection and kinetic simulation were conducted to rationally perform optimization. Amazingly, <i>Luminesensor</i> was proved to be as sensitive as to sense <b>natural light</b> and even <b>bioluminescence</b>. With this sensor, spatiotemporal control of cellular behavior, such as <b>high-resolution 2D</b> and <b>3D bio-printing</b> using dim light and even <b>luminescence of iPad</b> were shown to be very easy. What’s more, we successfully implemented <b>cell-cell signaling using light</b>, which is the very first time in synthetic biology and of great importance for biotechnological use. | ||
</p> | </p> | ||
| Line 838: | Line 858: | ||
<div class="floatR"><img src="/wiki/images/b/b6/Peking2012_PhototaxisSPECSdot.gif" alt="" style="width:200px;"/></div> | <div class="floatR"><img src="/wiki/images/b/b6/Peking2012_PhototaxisSPECSdot.gif" alt="" style="width:200px;"/></div> | ||
<div class="floatC"><p class="context_block" style="width:360px;"> | <div class="floatC"><p class="context_block" style="width:360px;"> | ||
| - | We | + | We did an awesome modeling to guide the optimization of our Luminesensor, combining protein kinetics, thermodynamics, and stochastic simulation with molecular docking together. Thereby the mutagenesis is indeed rationally guided by our modeling result! On the other hand, when we were modeling the process of photo-taxis, we also developed a hexagonal-coordinate simulation environment for dynamic system on a continuous plane, which would be a very useful prototype for future simulation on cellular movement, pattern formation and many other potential systems. |
</p></div> | </p></div> | ||
</div> | </div> | ||
Revision as of 14:10, 21 September 2012
Abstract |
HERE HERE
O
ptogenetic tools have made significant impact on life sciences and beyond. However, several serious issues remain: cytotoxicy, narrow dynamic range, and dependency on laser and exogenous chromophore. To circumvent these, Peking iGEM has rationally constructed a hypersensitive sensor of luminance -- Luminesensor. Primarily, the sensor was designed by fusing blue-light-sensing protein domain from Neurospora with DNA binding domain of LexA from E. coli, following which protein structure inspection and kinetic simulation were conducted to rationally perform optimization. Amazingly, Luminesensor was proved to be as sensitive as to sense natural light and even bioluminescence. With this sensor, spatiotemporal control of cellular behavior, such as high-resolution 2D and 3D bio-printing using dim light and even luminescence of iPad were shown to be very easy. What’s more, we successfully implemented cell-cell signaling using light, which is the very first time in synthetic biology and of great importance for biotechnological use.
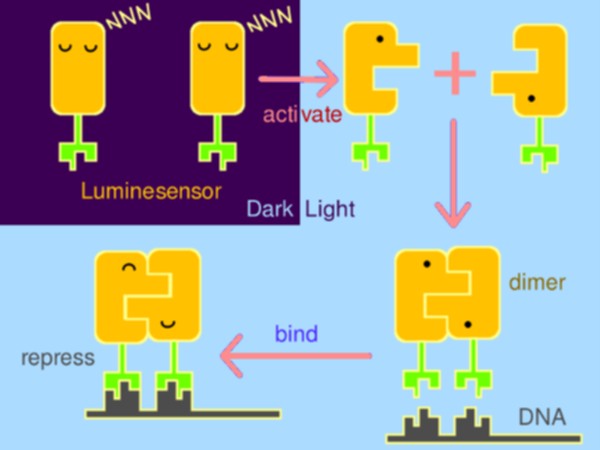
With strong motivation to raise a new generation of optogenetics, Peking iGEM team has rationally constructed a hypersensitive sensor of luminance – what we call the Luminesensor, with which spatiotemporal control of biochemical process or cellular behavior is highly feasible. It provides a paradigm for rational design of sensor.

The ultrasensitive Luminesensor is able to respond to very dim light and still maintain a high dynamic range. That encouraged Peking iGEM to explore the possibility of cell-cell communication through light. We have successfully implemented that , for the very first time, light-communication among cells without direct physical contact.

3D printing is a new technology that has been rising for many years. But in the realm of synthetic biology, it is far from developed. We exploited our Luminesensor to implement 3D printing that can be utilized in many applications in medical or manufacturing.
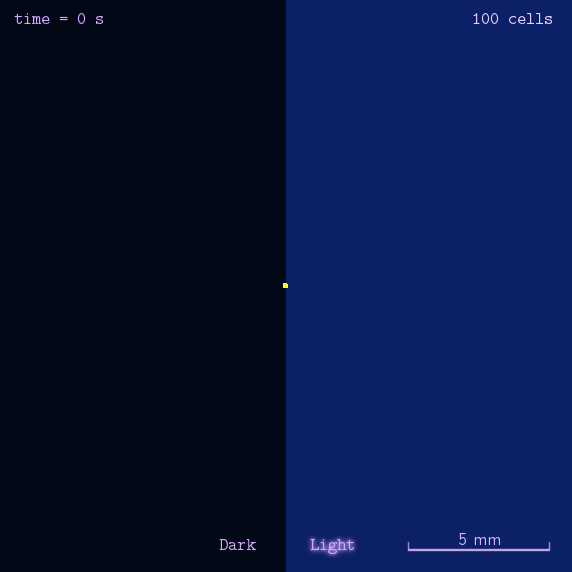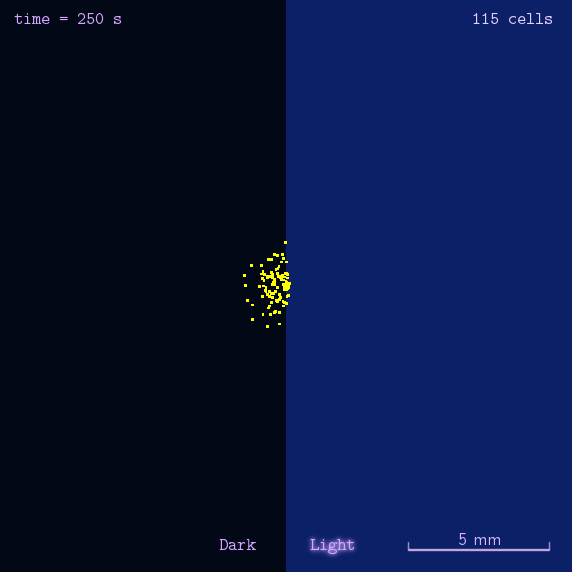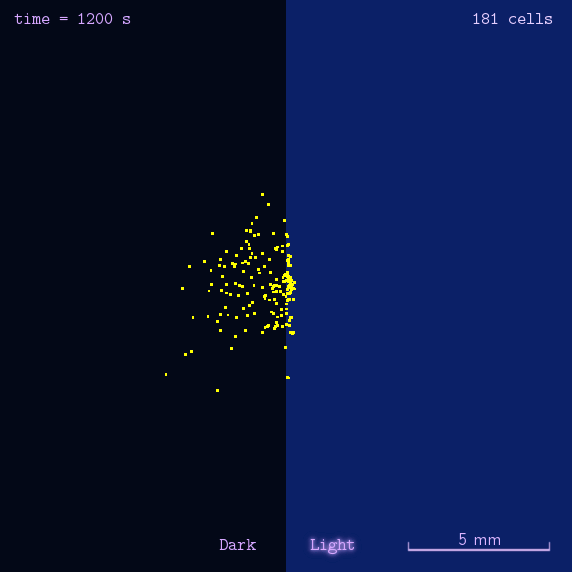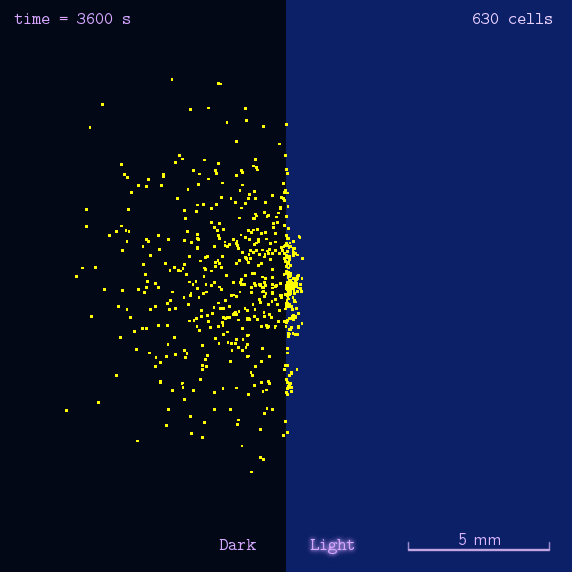
"Phototatic" bacteria can be built by programming the chemotaxis system in E. coli through light. By controlling the expression level of the cheZ protein with Luminesensor, the tumbling frequency is coupled to the intensity of light signals.

We have done a remarkable job in motivating high school students to study synthetic biology and guiding them towards future participation in the iGEM high school division.

We did an awesome modeling to guide the optimization of our Luminesensor, combining protein kinetics, thermodynamics, and stochastic simulation with molecular docking together. Thereby the mutagenesis is indeed rationally guided by our modeling result! On the other hand, when we were modeling the process of photo-taxis, we also developed a hexagonal-coordinate simulation environment for dynamic system on a continuous plane, which would be a very useful prototype for future simulation on cellular movement, pattern formation and many other potential systems.
 "
"















