Team:NYU Gallatin/Modeling
From 2012.igem.org
(Difference between revisions)
m |
|||
| (5 intermediate revisions not shown) | |||
| Line 45: | Line 45: | ||
<div class="content clearfix"> | <div class="content clearfix"> | ||
| - | <div class="field field-name-body field-type-text-with-summary field-label-hidden"><div class="field-items"><div class="field-item even">< | + | <div class="field field-name-body field-type-text-with-summary field-label-hidden"><div class="field-items"><div class="field-item even"><h1>Bio Synthetic Pathways</h1> |
| - | </div></div></div> </div> | + | <p></p><center><img src="http://farm9.staticflickr.com/8175/8052145533_c321d3ac67_z.jpg" /></center> |
| + | <h1>Codonizer</h1> | ||
| + | <p>To ensure that our modeling worked, we ran our sequences through a custom developed codon optimizer. Try it our yourself below! Drop in a chunk of DNA data and click "Optimize Codons" to have your sequence optimized for the acetobacter preferred codons.</p> | ||
| + | <p><textarea name="codons" id="old-codons" rows="10" cols="80"></textarea><input type="button" id="optimize-codons" value="Optimize Codons" /></p> | ||
| + | <div id="new-codons"></div> | ||
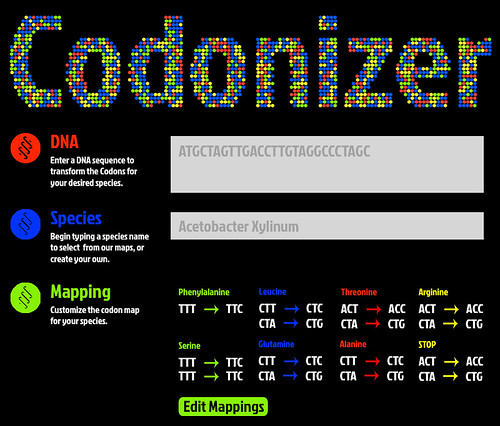
| + | <p>Upon completion of the iGEM competition we would like to expand our Codonizer software to support other strains. A mockup of what this tools will look like is provided below.</p> | ||
| + | <p></p><center><img src="http://farm9.staticflickr.com/8460/8052465891_a67ab6b567.jpg" /></center> | ||
| + | <script type="text/javascript" src="https://static.igem.org/mediawiki/2012/2/2e/Aseatobacter-Javascript_Codonizer.txt"></script></div></div></div> </div> | ||
Latest revision as of 03:09, 4 October 2012
 "
"