Team:NRP-UEA-Norwich/Week8
From 2012.igem.org
(→Labs) |
Khadijaouadi (Talk | contribs) |
||
| (One intermediate revision not shown) | |||
| Line 5: | Line 5: | ||
=Week 8= | =Week 8= | ||
| - | This has been the week of restriction digests. We have been working hard on trying to get fluorescent proteins ready for | + | This has been the week of restriction digests. We have been working hard on trying to get fluorescent proteins ready for ligation into our BioBricks. Three BioBricks have also been prepared for sequencing. We're hopeful that the sequenced confirms that the samples are in order with our planned BioBricks. We also finished the statistical analysis of the results of our growth study carried out a couple of weeks ago. It can be found on the [https://2012.igem.org/Team:NRP-UEA-Norwich/Experiments here]. |
| Line 11: | Line 11: | ||
===Lab=== | ===Lab=== | ||
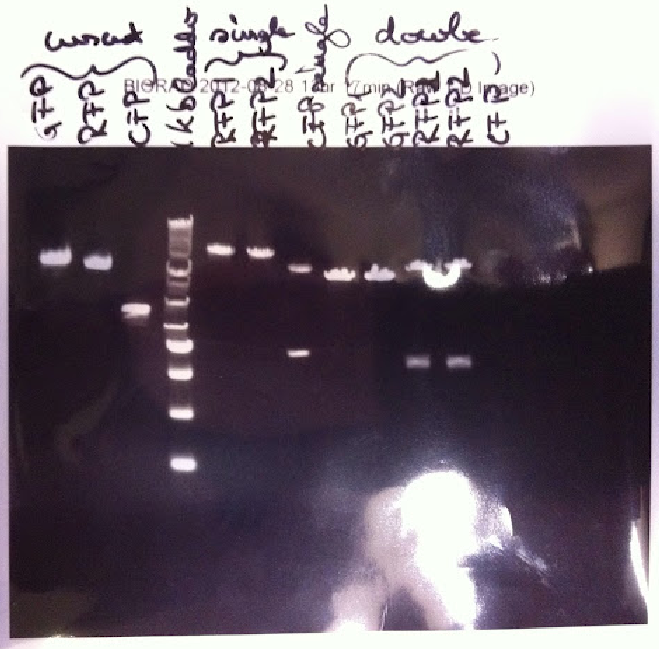
| - | [[File:27.jpg | thumb | '' '''Figure 1.''' Plasmid preps of CFP, GFP, RFP | + | [[File:27.jpg | thumb | '' '''Figure 1.''' Plasmid preps of CFP, GFP, RFP BioBricks, single digested with PstI, double digested using PstI and XbaI'']] |
| - | Khadija carried out a double digest on CFP, GFP and RFP. | + | Khadija carried out a double digest on CFP, GFP and RFP. The samples were digested with PstI and XbaI and left overnight. The mastermix was prepared using 10µl Buffer H, 1µl BSA, 2.5µl XbaI and 2.5µl PstI. This was evenly distributed into 5 tubes. In addition the final experimental solutions contained: 3µl GFP DNA and 13.8µl of water (nuclease free), 3.94 µl GFP DNA and 12.86 µl water, 3.47 µl RFP NDA and 13.33 µl water, 3.59 µl RFP DNA and 13.21 µl water and 3.6 µl CFP DNA and 13.2 µl of water. |
| - | The single digests from the previous Friday were run on a 1% agarose gel. The gel showed that GFP was cut once and linearised, as expected | + | The single digests from the previous Friday were run on a 1% agarose gel. The gel showed that GFP was cut once and linearised, as expected when cut by either PstI or EcoRV. The fragment length was estimated to be around 3000 bp. If in theory the linearised fragment was actually GFP, it would have a size of around 2800 bp. The digest of the RFP BioBrick produced two fragments. This should have not occurred. We decided to repeat the single digest of RFP. |
| - | The single digest of RFP was carried out alongside the single digest of CFP. We decided | + | The single digest of RFP was carried out alongside the single digest of CFP. We decided to only single digests with Pst1 to validate. The quantities remained the same as in the previous attempt of the experiment with 2µl Buffer H, 0.2µl BSA, 0.5µl, 1 µl DNA and 16.3 µl of water. The samples were left overnight to digest (both the single and double digests) (''Figure 1.''). |
| - | Looking at the plates from Friday which had been stored in the fridge after a day in an incubator, we | + | Looking at the plates from Friday, which had been stored in the fridge after a day in an incubator, we observed viable cells on all plates. Both the negative and positive controls showed the expected level of growth. We inoculated LB broth with cells from the plates containing colonies transformed with the BBa_K561002 (PFDHF +RFP + TETR). These we left in a shaking incubator overnight at 37degrees. In total three colonies were inoculated into three 5 ml of LB with chloramphenicol. |
==Day 2 (28/08/12)== | ==Day 2 (28/08/12)== | ||
| Line 29: | Line 29: | ||
===Lab=== | ===Lab=== | ||
| - | A 1% agarose gel was | + | A 1% agarose gel was prepared by Rebecca and poured whilst Khadija began setting up the completed restriction digest products to be run in a gel electrophoresis. 15µl of the single digests were mixed with 5µl of loading dye whilst 20µl of double digested DNA was added to 3µl of loading dye. The wells were loaded with the prepared samples and the electrophoresis took place for 1hour and 30mins. |
| - | After | + | After analysing the gel, we noticed the bands to be not straight. We then observed that the gel itself had not set straight, therefore we suspected the inconsistent thickness of the gel to have affected the outcome. We found that whilst the cut out fragments were what we expected them to be, at around 800bp, the plasmid backbone was evenly 1000bp out. We decided to repeat the digests. |
| - | The single digest was completely redone. As originally multiple colonies were inoculated, we decided to digest all the samples we had. In total we | + | The single digest was completely redone. As originally multiple colonies were inoculated, we decided to digest all the samples we had. In total we used 11 samples (3 GFP, 5 RFP and 4 CFP). Again the quantities in each solution were as followed: 2µl Buffer H, 0.2µl BSA, 0.5µl, 1 µl DNA and 16.3 µl of water. These were digested overnight. |
The grown overnight cultures of PFDHF + RFP + TETR were miniprepped. The concentrations of these have yet to be determined. | The grown overnight cultures of PFDHF + RFP + TETR were miniprepped. The concentrations of these have yet to be determined. | ||
| Line 42: | Line 42: | ||
===Lab=== | ===Lab=== | ||
| - | A 1% agarose gel was made and the overnight single digests of CFP, RFP and GFP were run on | + | A 1% agarose gel was made and the overnight single digests of CFP, RFP and GFP were run on an agarose gel. The samples were loaded alongside their uncut versions. The gel confirmed GFP being the BioBrick we were trying to purify. The other bricks showed different lengths, therefore we were certain that they were not of desired sequence. |
| - | Following this the GFP samples | + | Following this, the validated GFP samples underwent a double digest using XbaI and PstI. The double digest was utilised to separate the GFP insert from the plasmid backbone so that it can be ligate into our Bioricks MB and BM with the function of a reporter. The GFP samples that we used were of the following concentrations: GFP1 = 332.9ng/µl, GFP2 = 253.7ng/µl, GFP3 = 255.8ng/µl and GFP4 = 267.1 ng/µl. Using this data 3µl of GFP1, 3.94µl of GFP2, 3.9µl of GFP3 and 3.79µl of GFP4 was mixed with 3.2µl of a master mix solution consisting of 2.5µl of Buffer H, 1µl BSA, 2.5µl XbaI and 2.5µl PstI. Water was added to each of these to make each sample up to 20µl. To incubate they were left overnight. |
Following yesterday's minipreps of PFDHF + RFP + TETR, we nanodropped the isolated plasmids. The concentrations were very high. With 3 inoculations made from each colony, these repeats of each colony were labelled a, b and c. The concentrations were as follows: | Following yesterday's minipreps of PFDHF + RFP + TETR, we nanodropped the isolated plasmids. The concentrations were very high. With 3 inoculations made from each colony, these repeats of each colony were labelled a, b and c. The concentrations were as follows: | ||
| Line 60: | Line 60: | ||
3c) 522.4ng/µl | 3c) 522.4ng/µl | ||
| - | As we | + | As we were low on the pSB1C3 + MB/BM bricks that we wanted to send for sequencing and to iGEM, we ran single and double digests of these. The single digests were to validate existing BM/MB and the double digest was to take out the small fragment between Spe1 and Pst1 at the suffix to ligate the reporter GFP into. The single digest involved the following smaples: BM1, BM2, BM3, MB1, MB2, MB3 and MB4. In each of these digests 2µl DNA, 0.5µl PstI, 0.2 BSA, 2µl Buffer H were mixed and water was added to result in a final sample volume of 20µl. The double digest involved 1ng of DNA (this was based on the concentrations of the minipreps obtained from nanodropping). To these 0.5 XbaI , 0.5µl SpeI, 0.2 BSA, 2µl Buffer H and water was added to make each sample up to 20µl. |
==Day 4 (30/08/12)== | ==Day 4 (30/08/12)== | ||
===Labs=== | ===Labs=== | ||
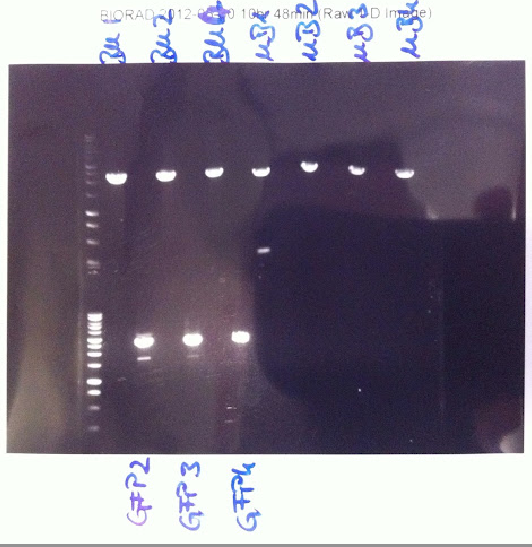
| - | [[File:30 blue.png | thumb | '' '''Figure 2.''' Double digest of B-M/M-B | + | [[File:30 blue.png | thumb | '' '''Figure 2.''' Double digest of B-M/M-B BioBricks with SpeI and PstI, double digest of GFP BioBrick with XbaI and PstI'' ]] |
| - | Lukas and Joy ran the double | + | Lukas and Joy ran the double and the single digests planned yesterday. The double digests of GFP showed that the DNA isolated was not GFP. Despite there being two bands, the fragments were too large. The single and double digests of BM and MB showed positive results. There were single clear bands at the expected size distance. These were excised from the gel and purified (''Firgure 2.'') |
| - | + | The new minipreps of BM and MB were packaged and prepared to be sent off to iGEM. The same DNA was sent for sequencing. | |
| - | + | As GFP, RFP and CFP that we previously had worked with, were not what we expected them to be, we decided to start afresh with new reporter BioBricks. On our plates we found BBa_E0040 and BBa_E1010 which are also GFP and RFP, with RBS sites respectively. We transformed these into Bioline alpha competent cells. Two controls were used: negative control of competent cells and a positive control of alpha competent cells transformed with PR8209 which also contains kanamycin resistance that it is shared with the two new BioBricks. We performed two separate transformations in order to save time and to minimise the danger of loosing DNA in case the transformation was faulty. Rachel and Pascoe performed one set of transformations and Rebecca and Khadija did another. | |
| - | + | ||
| - | As GFP, RFP and CFP that we previously had worked with were not what we expected them to be, we decided to start afresh with new reporter BioBricks. On our plates we found BBa_E0040 and BBa_E1010 which are also GFP and RFP | + | |
==Day 5 (31/08/12)== | ==Day 5 (31/08/12)== | ||
| Line 79: | Line 77: | ||
===Dry Lab=== | ===Dry Lab=== | ||
| - | The | + | The sequence of our MB and BM BioBricks was returned to us and the analysis began. BM3 was found to have the sequence we expected it to have. We will send this sample to iGEM HQ and hopefully have officially made a BioBrick. Most of the sequences confirmed that they were what we expected them to be with the exceptions of the newly made series. Unfortunately, we do not have sufficient amounts of the other samples to send to iGEM. We are now looking into requesting them back from Bio Liefesciences. |
| - | Statistical analysis | + | Statistical analysis of the Growth Study data, carried out last week, showed that the growth rates between Alpha cells (Control) and PyeaR, MB and BM are not significantly different. However, the independent T Test significance level for PyeaR is 0.050 and hence, despite not being significantly different at the 95% confidence level, it is significantly different at the 90% confidence. There was also no significant difference between BM and MB. There was however, a significant difference between PyeaR transformed cells and BM and MB. |
Latest revision as of 00:36, 27 September 2012
 "
"