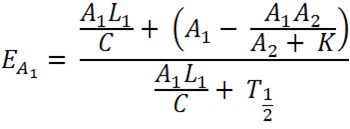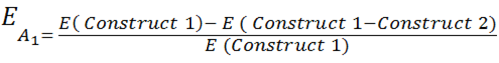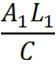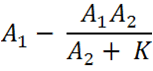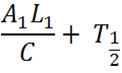Team:NRP-UEA-Norwich/Theoretical Characterisation
From 2012.igem.org
| (17 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
==Comparator Circuit Characterisation== | ==Comparator Circuit Characterisation== | ||
| - | The idea of the comparator circuit is to provide a modular sensor which can specifically and quantitatively measure different chemical species within the cell. Through | + | The idea of the comparator circuit is to provide a modular method of signal integration that can produce a sensor which can specifically and quantitatively measure different chemical species within the cell using non specific promoters. Through mathematical modelling, an equation has been assembled which can predict the expression of each of the reporter proteins such as RFP and CFP. |
[[File:Equation_7.png| 300px | center]] | [[File:Equation_7.png| 300px | center]] | ||
| - | Figure: Theoretical equation to | + | Figure: Theoretical equation to predict the degree of expression of Construct 1 and 2. |
| - | + | The full equation has been laid out in a way that is relevant only to Construct 1, however, the numbers can be reversed to be relevant to Construct 2. For ease of explanation, everything described will be relevant to Construct 1. | |
| - | E = Proportion of rate | + | E = Proportion of expression rate of Construct 1 when both constructs are transcribed (i.e. there is knockdown of one construct) relative to the non-knocked down expression of Construct 1 when only Construct 1 is expressed. |
| - | A = The rate of transcription of Construct 1 as a proportion of the maximum transcription rate. As a proportion this is measured on a scale of 0 - 1. As an example if the rate of transcription is half of the maximum rate, rate would be 0.5 (arbituary units). It can be assumed the rate of transcription of construct 1 and 2 due to cellular components (e.g. RNA polymerase) is the same, however, the | + | A = The rate of transcription of Construct 1 as a proportion of the maximum transcription rate. As a proportion this is measured on a scale of 0 - 1. As an example if the rate of transcription is half of the maximum rate, rate would be 0.5 (arbituary units). It can be assumed the rate of transcription of construct 1 and 2 due to cellular components (e.g. RNA polymerase) is the same, however, the rate of transcription initiation will dictate the transcription rate. The initiation is reliant on the chemical species interacting with the transcription factor which binds to the promoter (i.e. nitric oxide,nitrates,nitrites to PyeaR). The '1' and '2' refer to the Construct 1 or 2 and hence the promoter and the measured fluorescent protein attached (e.g GFP, RFP, CFP, etc). |
L = The length of the Construct 1 in the DNA form that is transcribed (i.e the leader and protein coding region). | L = The length of the Construct 1 in the DNA form that is transcribed (i.e the leader and protein coding region). | ||
| - | Note: Leader refers to the section of RNA at the start of the mRNA that is not translated but has an | + | Note: Leader refers to the section of RNA at the start of the mRNA that is not translated but has an effect on translation rate. |
C = The rate of transcription. Assuming the rate of transcription of Construct 1 and 2 are the same because the same ribosomes and RBS are involved. | C = The rate of transcription. Assuming the rate of transcription of Construct 1 and 2 are the same because the same ribosomes and RBS are involved. | ||
| Line 26: | Line 26: | ||
K = A constant of the biological system. This can only be measured through observation. | K = A constant of the biological system. This can only be measured through observation. | ||
| - | |||
| - | [[File:Equation_2.png| | + | The full equation is modelled on the basic equation of: |
| + | |||
| + | [[File:Equation_2.png| 400px | center]] | ||
where E is the rate of expression and E(A1) is the same as that explained above. | where E is the rate of expression and E(A1) is the same as that explained above. | ||
| - | The additional complexity factors in less assumptions, and | + | |
| + | The additional complexity factors in less assumptions, and mimics a biological system, more closely. Below is a breakdown of the full equation. | ||
[[File:Equation_3.png| 50px | left]] | [[File:Equation_3.png| 50px | left]] | ||
| - | This refers to the | + | This refers to the number of Construct 1 RNA transcripts undergoing transcription at any one time. The length of DNA is particularly important when the chassis is bacterial. In bacteria, as there is no true nucleus, translation occurs simultaneously with transcription. Transcription affects the probability of interaction between construct 1 and 2 and therefore, they are less likely to be translated. As the measurement of fluorescence is the output directly related to the rate of translation, the overall equation measures translation, however, translation rate iis dependent on rate of transcription and degree of knockdown, and hence transcription is factored in here. L/C is the period of time taken for transcription to take place. It is the time in which translation can be initiated but it is unlikely that the two leaders will bind to one another |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[File:Equation_5.png| 150px | left]] | [[File:Equation_5.png| 150px | left]] | ||
| + | This part of the equation is the deduction of the knockdown of Construct 1 when there is Construct 2 expression and interaction. The biological constant, k, factors in that not all of construct 2 that is expressed will interact with construct 1 and vice versa. Hence, both exist despite construct 2 existing in small quantities. We believe that depending upon the assembly of the orientation of the two constructs within the plasmid, the interaction and hence the binding efficiency can be altered vastly. If the genes have opposite orientations, so that the termination sites are very close then the reduction of distance will increase the chances of interaction and hence make the sensory system more accurate. | ||
| + | [[File:Equation_6.png| 120px | left]] | ||
| - | + | This part of the equation encompasses the natural half life of Construct 1 when it alone is expressed (i.e. no expression of or interaction with Construct 2). As described before in the modelling from the basic equation, this is the lower part of the equation and puts it in perspective of Construct 1 and gives expression as a porportion of the maximum transcription. The half life is also Construct 1's half life. | |
| - | + | ||
| - | . | + | |
| - | . | + | So to bring it all together; the top half of the equation indicates the degree of translation of the RNA transcribed by the first promoter under any particular transcription rate of the two promoters in arbitrary units. To make this into a meaningful output it is divided by the maximum translation rate at that rate of transcription to equal E(A1); this indicates the degree of attenuation of one RNA from the other. |
Latest revision as of 23:57, 23 September 2012
 "
"