Team:Macquarie Australia/ProjectOverview
From 2012.igem.org
Rationale
Being able to induce gene expression is one of the most important aspect of the biomolecular sciences. It allows us to determine the role of specific genes and to produce them in vast quantities. Chemical induction is commonly used but has some lag time and so is less useful to determine if the role of a developmental gene. As such a fast and reversible method of controlling gene expression needs to be developed, one that does not require a long incubation time. Our project aims to produce a light controlled gene switch by coupling heme oxygenase with a bacteriophytochrome. To achieve this goal we will be using Gibson Assembly.
The bacteriophytochrome requires biliverdin to function which will be supplied by heme oxygenase. When irradiated with red light the bacteriophytochrome undergoes a conformation change and metabolic pathways are activated.
To truly embrace synthetic biology and modify genes to suit our need we have utilised Gibson Assembly. This allows us to introduce numerous changes to the gene of interest. Our project is a showcase for the power of Gibson Assembly in Synthetic Biology. By starting with an a standard and optimising it for E. coli we will show how simple Gibson Assembly is in altering the gene but maintaining function.
Project Workflow
- Design of Heme oxygenase and two bacteriophytochrome fragments
We selected three genes for our project, a single heme oxygenase and bacteriophytochromes from Deinococcus radiodurans and Agrobacterium tumefaciens. They were designed to remove any internal restriction sites without the need for site directed mutagenesis. The genes will also be codon optimised for E. coli. In this way a heme oxygenase gene will be produced that is optimised for E. coli and as such more amenable to synthetic biology experiments.
- Assembly of fragment sequences into BioBricks using Gibson Assembly
Using Gibson Assembly we will reassemble our genes insert them into the plasmid backbone. This removes the need for ligations and restriction digests. Allowing the production of complete BioBricks without the need for extra steps to get the gene into the destination plasmid.
- Transform in E. coli
By transforming in E. coli we can determine if the gene is functional as well as purify the plasmid. By transforming in BL21 strain E. coli we can overproduce the protein and then characterise the BioBricks produced.
- Sequence clones
It is imperative that the plasmids produced from the Gibson Assembly be sequenced to determine if there have been any nucleotide changes between the planned sequences and those synthesised. Therefore sequencing data needs to be gathered before any ligations are performed to produce our self assembling switch. This will also demonstrate that the protein sequence has not changed and the protein should therefore be functional.
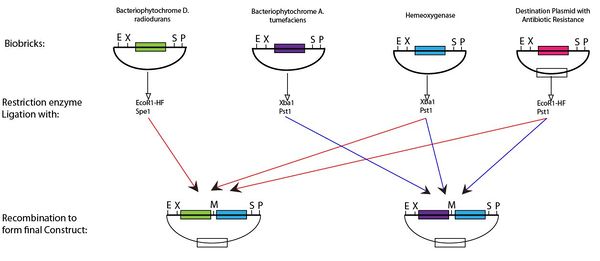
- Assemble BioBricks part to produce the gene switch
Following digestion of the BioBricks produced with the appropriate enzyme and ligation it is possible to produce the plasmid for the light switch. This protocol can be seen below,
- Transform and Characterise the light switch
After ligating the two BioBricks to assemble the light switch we will be able to show the usefulness of Gibson Assembly in synthetic biology. This will provide a means to characterise the two biobricks simultaneously.
- Assembly of fragment sequences into BioBricks using Gibson Assembly
 "
"