Team:Macquarie Australia/Project
From 2012.igem.org
(→Project Work Flow) |
|||
| Line 32: | Line 32: | ||
5. Sequence | 5. Sequence | ||
<br/> | <br/> | ||
| - | 6. Assemble | + | 6. Assemble BioBricks according to NEB & Gingko Bioworks Protocol |
<br/> | <br/> | ||
7. Transform into E. coli | 7. Transform into E. coli | ||
| Line 38: | Line 38: | ||
8. Test for function and sequence (26th September: Parts must be sent to registry by this date) | 8. Test for function and sequence (26th September: Parts must be sent to registry by this date) | ||
<br/> | <br/> | ||
| - | |||
| - | |||
| - | |||
| - | |||
== Results == | == Results == | ||
Revision as of 02:03, 4 September 2012
Overall project
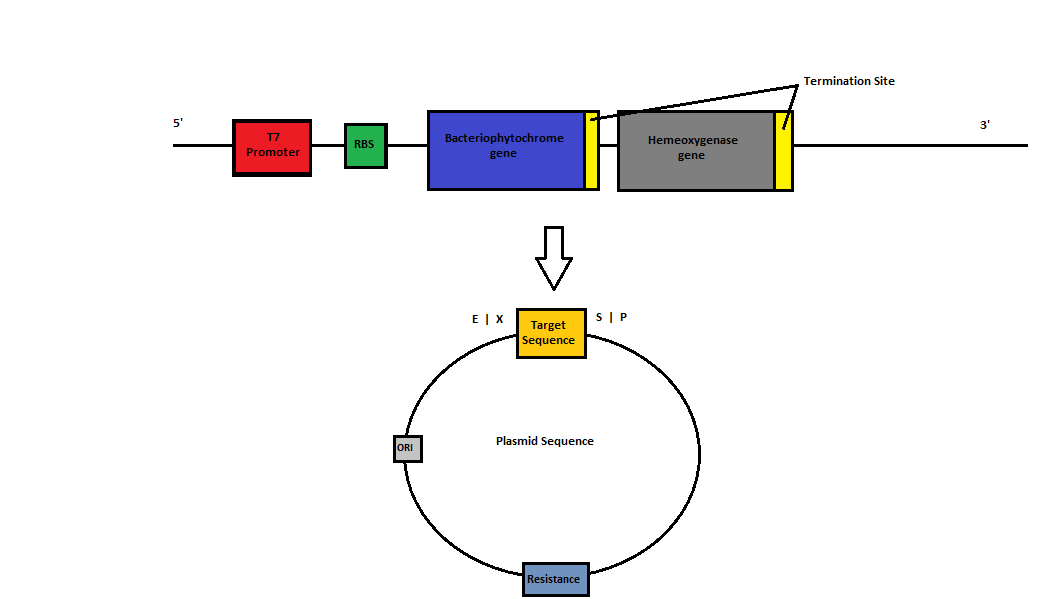
Background: Phytochromes are ubiquitous proteins that allow an organism to sense light. These proteins have evolved in unique environments to sense light intensity in different colour ranges. This experiment focuses on constructing a biological switch that uses phytochromes from Deinococcus radiodurans and Agrobacterium tumefaciens. The coupling of heme oxygenase supplies our phytochrome proteins with biliverdin, allowing for the self-assembly of the switch within host systems. The switch is the first stage of a two component light sensor and when expressed at high level, there is a noticeable colour change of the cell when it is activated by light.


- Bacteriophytochrome
- Hemeoxygenase
- Bacteriophytochrome
Project Aims: The objective of this project is to therefore build and characterise a biological light switch in E. coli. This will involve construction of heme-oxygenase and bacteriophytochrome BioBrick parts. This year's research team will be expanding upon the research conducted by last year's iGEM team and the team from 2010. In 2010 the Macquarie Team cloned bacteriophytochrome from two sources. They showed that when one was expressed, it was functionally assembled when incubated with exogenous biliverdin and able to elicit a colour change when excited with far-red light. However, the part created is not directly usable as a BioBrick as it contains an internal EcoRI site (Deinococcus radiodurans phytochrome) and 2 PstI sites (Agrobacterium tumefaciens phytochrome). As biliverdin is not native to E. coli, the addition of heme oxygenase is required for the synthesis of bilivedin, enabling the self-assembly of the light switch. In 2011, the Macquarie Team successfully managed to construct and characterise the heme oxygenase 1 as a BioBrick. They showed, via its green color, that cells expressing the heme oxygenase could degrade heme into biliverdin.
In this project we will aim to construct the BioBricks using Gibson cloning as opposed to restriction enzyme digests.
Project Work Flow
1. Design of Heme oxygenase and two bacteriophytochrome fragments
2. Approval and synthesis of fragments by Integrated DNA Technologies (IDT)
3. Assembly of fragment sequences into BioBricks using Gibson Assembly
4. Transform into E. coli
5. Sequence
6. Assemble BioBricks according to NEB & Gingko Bioworks Protocol
7. Transform into E. coli
8. Test for function and sequence (26th September: Parts must be sent to registry by this date)
 "
"