Team:Macquarie Australia/Project
From 2012.igem.org
(→Project Work Flow) |
|||
| (26 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Macquarie_Australia/Template/MQ12}} | {{:Team:Macquarie_Australia/Template/MQ12}} | ||
| + | <html> | ||
| + | <center><h1>Overall Project</h1></center> | ||
| + | <h2>Rationale</h2> | ||
| + | <p>Being able to induce gene expression is one of the most important aspect of the biomolecular sciences. It allows us to determine the role of specific genes and to produce them in vast quantities. Chemical induction is commonly used but has some lag time and so is less useful to determine if the role of a developmental gene. As such a fast and reversible method of controlling gene expression needs to be developed, one that does not require a long incubation time. Our project aims to produce a light controlled gene switch by coupling heme oxygenase with a bacteriophytochrome. To achieve this goal we will be using Gibson Assembly.</p> | ||
| + | <p>The bacteriophytochrome requires biliverdin to function which will be supplied by heme oxygenase. When irradiated with red light the bacteriophytochrome undergoes a conformation change and metabolic pathways are activated.</p> | ||
| + | <p>To truly embrace synthetic biology and modify genes to suit our need we have utilised Gibson Assembly. This allows us to introduce numerous changes to the gene of interest. Our project is a showcase for the power of Gibson Assembly in Synthetic Biology. By starting with an a standard and optimising it for <i>E. coli</i> we will show how simple Gibson Assembly is in altering the gene but maintaining function.</p> | ||
| + | <br> | ||
| + | <h2>Relationship to Previous Teams' work</h2> | ||
| + | <p>Previous Macquarie Teams focused on coupling heme oxygenase with a bacteriophytochrome. They had little success in producing a functional switch. They did produce a BioBricks for the different components that contained illegal restriction site. Our project utilises the novel approach of Gibson Assembly to remove the need to perform traditional methods, such as PCR. We will not be using any parts produced or genomic DNA used by the previous Macquarie Teams. This team will produce codon optimised switch capable of controlling gene regulation for <i>E. coli</i> as well as removing restrictions sites that complicated ligation procedures.</p> | ||
| + | <h2>Abstract</h2> | ||
| + | <p>Phytochromes, or photoreceptors with the ability to control the expression of genes, exist in bacteria as bacteriophytochromes. This project creates a light-dependent biological switch using the bacteriophytochromes from <i>Deinococcus radiodurans</i> and <i>Agrobacterium tumefaciens</i>. When coupled with heme oxygenase, these bacteriophytochromes are supplied with biliverdin, a pigment which allows for the self-assembly of a switch within the host system. In the presence of red light, the conformation of the bacteriophytochrome is modified. This reaction produces a visible colour change in the presence of red light, and can be used to control expression of a targeted gene when coupled with the appropriate response regulator. Exposure to far-red light will cause the bacteriophytochrome to revert to its original conformation, thus repressing the gene and reversing the colour change.</p> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/f/f6/Bacteriophytochrome.gif"> <img src="https://static.igem.org/mediawiki/2012/7/79/Hemeoxygenase.gif"> </center> | ||
| + | <center> <i>Crystal Structures of Bacteriophytochrome and Heme Oxygenase</i></center> | ||
| + | <br> | ||
| + | <h2>Project Aims</h2> | ||
| + | <p>The objective of this project is to therefore build and characterise a biological light switch in <i>E. coli</i>. This will involve construction of heme-oxygenase and bacteriophytochrome BioBrick parts. This year's research team will be expanding upon the research conducted by last year's iGEM team and the team from 2010. In 2010 the Macquarie Team cloned bacteriophytochrome from two sources. They showed that when one was expressed, it was functionally assembled when incubated with exogenous biliverdin and able to elicit a colour change when excited with far-red light. However, the part created is not directly usable as a BioBrick as it contains an internal <i>Eco</i>RI site (<i>Deinococcus radiodurans</i> phytochrome) and 2 <i>Pst</i>I sites (<i>Agrobacterium tumefaciens</i> phytochrome). As biliverdin is not native to <i>E. coli</i>, the addition of heme oxygenase is required for the synthesis of bilivedin, enabling the self-assembly of the light switch. In 2011, the Macquarie Team successfully managed to construct and characterise the heme oxygenase 1 as a BioBrick. They showed, via its green colour, that cells expressing the heme oxygenase could degrade heme into biliverdin.</p> | ||
| - | + | <a name="flow"><h2>Project Workflow</h2></a> | |
| - | + | <ol> | |
| - | + | <a name="1"><h3><li>Design of Heme oxygenase and two bacteriophytochrome fragments</h3> | |
| - | + | <p>We selected three genes for our project, a single heme oxygenase and bacteriophytochromes from <i>Deinococcus radiodurans</i> and <i>Agrobacterium tumefaciens</i>. They were designed to remove any internal restriction sites without the need for site directed mutagenesis. The genes will also be codon optimised for <i>E. coli</i>. In this way a heme oxygenase gene will be produced that is optimised for <i>E. coli</i> and as such more amenable to synthetic biology experiments.</p> | |
| - | + | <br> | |
| - | < | + | <a name="2"><h3><li>Assembly of fragment sequences into BioBricks using Gibson Assembly</h3> |
| - | + | <p>Using Gibson Assembly we will reassemble our genes insert them into the plasmid backbone. This removes the need for ligations and restriction digests. Allowing the production of complete BioBricks without the need for extra steps to get the gene into the destination plasmid.</p> | |
| - | + | <br> | |
| - | + | <a name="3"><h3><li>Transform in <i>E. coli</i></h3> | |
| - | + | <p>By transforming in <i>E. coli</i> we can determine if the gene is functional as well as purify the plasmid. By transforming in BL21 strain <i>E. coli</i> we can overproduce the protein and then characterise the BioBricks produced.</p> | |
| - | + | <br> | |
| - | + | <a name="4"><h3><li>Sequence clones</h3> | |
| - | + | <p>It is imperative that the plasmids produced from the Gibson Assembly be sequenced to determine if there have been any nucleotide changes between the planned sequences and those synthesised. Therefore sequencing data needs to be gathered before any ligations are performed to produce our self assembling switch. This will also demonstrate that the protein sequence has not changed and the protein should therefore be functional.</p> | |
| - | + | <br> | |
| - | + | <a name="5"><h3><li>Assemble BioBricks part to produce the gene switch</h3> | |
| - | + | <p>Following digestion of the BioBricks produced with the appropriate enzyme and ligation it is possible to produce the plasmid for the light switch. This protocol can be seen below,</p> | |
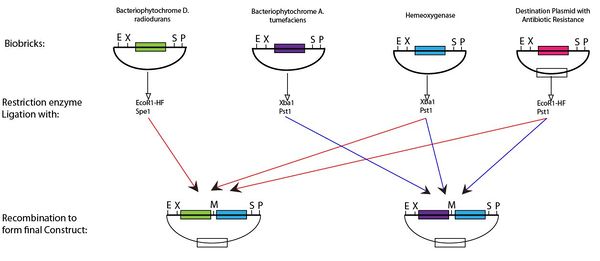
| - | + | <div class="center"><a name="5"></a><div class="thumb tnone"><a name="5"></a><div class="thumbinner" style="width:602px;"><a name="5"></a><a href="/File:Construct1111.JPG" class="image"><img alt="" src="/wiki/images/thumb/d/d9/Construct1111.JPG/600px-Construct1111.JPG" width="600" height="261" class="thumbimage"></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Construct1111.JPG" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt=""></a></div>Assembly of BioBricks via restriction enzyme digestion</div></div></div></div> | |
| - | == | + | <br> |
| - | < | + | <a name="6"><h3><li>Transform and Characterise the light switch</h3> |
| - | + | <p>After ligating the two BioBricks to assemble the light switch we will be able to show the usefulness of Gibson Assembly in synthetic biology. This will provide a means to characterise the two biobricks simultaneously.</p></ol><br> | |
| - | + | <p>Our workflow is expressed below in the form of a flow chart. To go into greater depth with that particular section simply click on the box.</p> | |
| - | + | <br> | |
| - | < | + | <html><center><img src="https://static.igem.org/mediawiki/2012/b/b8/MQMAP2.png" usemap="#MQ" border="0" width ="750" height="375"> |
| - | + | <map name="MQ"> | |
| - | < | + | <area shape="rect" coords="35,18,127,70" href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/GibsonTips" /> |
| - | + | <area shape="rect" coords="61,100,168,151" href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/ArrivalofGBlocks" /> | |
| - | < | + | <area shape="rect" coords="226,100,332,151" href="https://2012.igem.org/Team:Macquarie_Australia/Results" /> |
| - | + | <area shape="rect" coords="74,249,143,303" href="https://2012.igem.org/Team:Macquarie_Australia/Results#1" /> | |
| - | < | + | <area shape="rect" coords="197,262,265,316" href="https://2012.igem.org/Team:Macquarie_Australia/Results#3" /> |
| - | + | <area shape="rect" coords="224,194,293,248" href="https://2012.igem.org/Team:Macquarie_Australia/Results#3" /> | |
| - | + | <area shape="rect" coords="37,185,200,228" href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/GibsonAssembly" /> | |
| - | + | <area shape="rect" coords="417,263,525,343" href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/ligations" /> | |
| - | + | <area shape="rect" coords="388,16,553,221" href="https://2012.igem.org/Team:Macquarie_Australia/Results#7"/> | |
| - | + | <area shape="rect" coords="609,139,730,193" href="https://2012.igem.org/Team:Macquarie/Protocols/TransformationProtocol" /> | |
| - | + | <area shape="rect" coords="607,235,731,274" href="https://2012.igem.org/Team:Macquarie_Australia/Results#7" /> | |
| - | < | + | </map> |
| - | + | </map> | |
| - | < | + | </center> |
| - | + | <p> | |
| - | < | + | <h2>Significance of the Project</h2> |
| - | + | </p> | |
| - | + | <p> | |
| - | < | + | <b>The success of our project allows the construction of a reversible, inexpensive and non-invasive mechanism of gene regulation</b>. This is a valuable tool for research and industrial practice in a number of scientific fields, including biomolecular science, nutrition, medicine and agriculture. Our project offers researchers a model for refining and employing current methods of gene regulation. |
| - | + | </p> | |
| - | + | </html> | |
Latest revision as of 00:11, 27 September 2012
Overall Project
Rationale
Being able to induce gene expression is one of the most important aspect of the biomolecular sciences. It allows us to determine the role of specific genes and to produce them in vast quantities. Chemical induction is commonly used but has some lag time and so is less useful to determine if the role of a developmental gene. As such a fast and reversible method of controlling gene expression needs to be developed, one that does not require a long incubation time. Our project aims to produce a light controlled gene switch by coupling heme oxygenase with a bacteriophytochrome. To achieve this goal we will be using Gibson Assembly.
The bacteriophytochrome requires biliverdin to function which will be supplied by heme oxygenase. When irradiated with red light the bacteriophytochrome undergoes a conformation change and metabolic pathways are activated.
To truly embrace synthetic biology and modify genes to suit our need we have utilised Gibson Assembly. This allows us to introduce numerous changes to the gene of interest. Our project is a showcase for the power of Gibson Assembly in Synthetic Biology. By starting with an a standard and optimising it for E. coli we will show how simple Gibson Assembly is in altering the gene but maintaining function.
Relationship to Previous Teams' work
Previous Macquarie Teams focused on coupling heme oxygenase with a bacteriophytochrome. They had little success in producing a functional switch. They did produce a BioBricks for the different components that contained illegal restriction site. Our project utilises the novel approach of Gibson Assembly to remove the need to perform traditional methods, such as PCR. We will not be using any parts produced or genomic DNA used by the previous Macquarie Teams. This team will produce codon optimised switch capable of controlling gene regulation for E. coli as well as removing restrictions sites that complicated ligation procedures.
Abstract
Phytochromes, or photoreceptors with the ability to control the expression of genes, exist in bacteria as bacteriophytochromes. This project creates a light-dependent biological switch using the bacteriophytochromes from Deinococcus radiodurans and Agrobacterium tumefaciens. When coupled with heme oxygenase, these bacteriophytochromes are supplied with biliverdin, a pigment which allows for the self-assembly of a switch within the host system. In the presence of red light, the conformation of the bacteriophytochrome is modified. This reaction produces a visible colour change in the presence of red light, and can be used to control expression of a targeted gene when coupled with the appropriate response regulator. Exposure to far-red light will cause the bacteriophytochrome to revert to its original conformation, thus repressing the gene and reversing the colour change.


Project Aims
The objective of this project is to therefore build and characterise a biological light switch in E. coli. This will involve construction of heme-oxygenase and bacteriophytochrome BioBrick parts. This year's research team will be expanding upon the research conducted by last year's iGEM team and the team from 2010. In 2010 the Macquarie Team cloned bacteriophytochrome from two sources. They showed that when one was expressed, it was functionally assembled when incubated with exogenous biliverdin and able to elicit a colour change when excited with far-red light. However, the part created is not directly usable as a BioBrick as it contains an internal EcoRI site (Deinococcus radiodurans phytochrome) and 2 PstI sites (Agrobacterium tumefaciens phytochrome). As biliverdin is not native to E. coli, the addition of heme oxygenase is required for the synthesis of bilivedin, enabling the self-assembly of the light switch. In 2011, the Macquarie Team successfully managed to construct and characterise the heme oxygenase 1 as a BioBrick. They showed, via its green colour, that cells expressing the heme oxygenase could degrade heme into biliverdin.
Project Workflow
- Design of Heme oxygenase and two bacteriophytochrome fragments
We selected three genes for our project, a single heme oxygenase and bacteriophytochromes from Deinococcus radiodurans and Agrobacterium tumefaciens. They were designed to remove any internal restriction sites without the need for site directed mutagenesis. The genes will also be codon optimised for E. coli. In this way a heme oxygenase gene will be produced that is optimised for E. coli and as such more amenable to synthetic biology experiments.
- Assembly of fragment sequences into BioBricks using Gibson Assembly
Using Gibson Assembly we will reassemble our genes insert them into the plasmid backbone. This removes the need for ligations and restriction digests. Allowing the production of complete BioBricks without the need for extra steps to get the gene into the destination plasmid.
- Transform in E. coli
By transforming in E. coli we can determine if the gene is functional as well as purify the plasmid. By transforming in BL21 strain E. coli we can overproduce the protein and then characterise the BioBricks produced.
- Sequence clones
It is imperative that the plasmids produced from the Gibson Assembly be sequenced to determine if there have been any nucleotide changes between the planned sequences and those synthesised. Therefore sequencing data needs to be gathered before any ligations are performed to produce our self assembling switch. This will also demonstrate that the protein sequence has not changed and the protein should therefore be functional.
- Assemble BioBricks part to produce the gene switch
Following digestion of the BioBricks produced with the appropriate enzyme and ligation it is possible to produce the plasmid for the light switch. This protocol can be seen below,
- Transform and Characterise the light switch
After ligating the two BioBricks to assemble the light switch we will be able to show the usefulness of Gibson Assembly in synthetic biology. This will provide a means to characterise the two biobricks simultaneously.
- Assembly of fragment sequences into BioBricks using Gibson Assembly
Our workflow is expressed below in the form of a flow chart. To go into greater depth with that particular section simply click on the box.

Significance of the Project
The success of our project allows the construction of a reversible, inexpensive and non-invasive mechanism of gene regulation. This is a valuable tool for research and industrial practice in a number of scientific fields, including biomolecular science, nutrition, medicine and agriculture. Our project offers researchers a model for refining and employing current methods of gene regulation.
 "
"