Team:Macquarie Australia/Project
From 2012.igem.org
| Line 19: | Line 19: | ||
<html><center><img src="https://static.igem.org/mediawiki/2012/b/b8/MQMAP2.png" usemap="#MQ" border="0" width ="750" height="375"> | <html><center><img src="https://static.igem.org/mediawiki/2012/b/b8/MQMAP2.png" usemap="#MQ" border="0" width ="750" height="375"> | ||
<map name="MQ"> | <map name="MQ"> | ||
| - | <area shape="rect" coords="35,18,127,70" href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/ | + | <area shape="rect" coords="35,18,127,70" href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/GibsonTips" /> |
<area shape="rect" coords="61,100,168,151" href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/ArrivalofGBlocks" /> | <area shape="rect" coords="61,100,168,151" href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/ArrivalofGBlocks" /> | ||
| - | <area shape="rect" coords="226,100,332,151" href="https://2012.igem.org/Team:Macquarie_Australia/Results | + | <area shape="rect" coords="226,100,332,151" href="https://2012.igem.org/Team:Macquarie_Australia/Results" /> |
| - | <area shape="rect" coords="74,249,143,303" href=" | + | <area shape="rect" coords="74,249,143,303" href="https://2012.igem.org/Team:Macquarie_Australia/Results#1" /> |
| - | <area shape="rect" coords="197,262,265,316" href=" | + | <area shape="rect" coords="197,262,265,316" href="https://2012.igem.org/Team:Macquarie_Australia/Results#3" /> |
| - | <area shape="rect" coords="224,194,293,248" href=" | + | <area shape="rect" coords="224,194,293,248" href="https://2012.igem.org/Team:Macquarie_Australia/Results#3" /> |
| - | <area shape="rect" coords="37,185,200,228" href=" | + | <area shape="rect" coords="37,185,200,228" href="NEED TO PUT IN THAT SPECIFIC PROTOCOL" /> |
<area shape="rect" coords="417,263,525,343" href="nohref" /> | <area shape="rect" coords="417,263,525,343" href="nohref" /> | ||
<area shape="rect" coords="388,16,553,221" href="nohref" /> | <area shape="rect" coords="388,16,553,221" href="nohref" /> | ||
Revision as of 04:24, 22 September 2012
Overall project
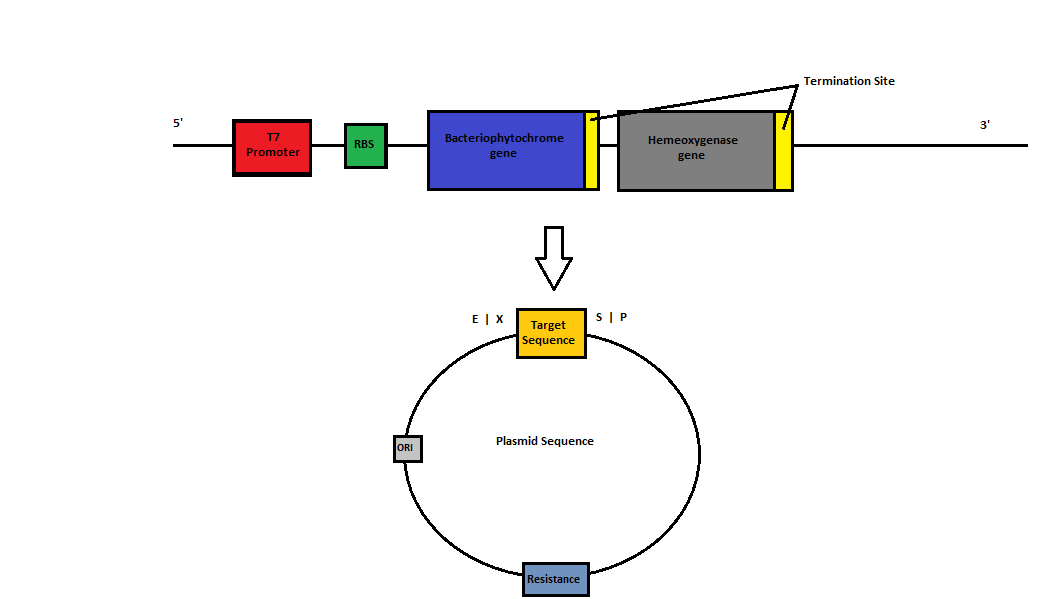
Abstract: Phytochromes, or photoreceptors with the ability to control the expression of genes, exist in bacteria as bacteriophytochromes. This project creates a light-dependent biological switch using the bacteriophytochromes from Deinococcus radiodurans and Agrobacterium tumefaciens. When coupled with heme oxygenase, these bacteriophytochromes are supplied with biliverdin, a pigment which allows for the self-assembly of a switch within the host system. In the presence of red light, the conformation of the bacteriophytochrome is modified. This reaction produces a visible colour change in the presence of red light, and can be used to control expression of a targeted gene when coupled with the appropriate response regulator. Exposure to far-red light will cause the bacteriophytochrome to revert to its original conformation, thus repressing the gene and reversing the colour change.


Project Aims: The objective of this project is to therefore build and characterise a biological light switch in E. coli. This will involve construction of heme-oxygenase and bacteriophytochrome BioBrick parts. This year's research team will be expanding upon the research conducted by last year's iGEM team and the team from 2010. In 2010 the Macquarie Team cloned bacteriophytochrome from two sources. They showed that when one was expressed, it was functionally assembled when incubated with exogenous biliverdin and able to elicit a colour change when excited with far-red light. However, the part created is not directly usable as a BioBrick as it contains an internal EcoRI site (Deinococcus radiodurans phytochrome) and 2 PstI sites (Agrobacterium tumefaciens phytochrome). As biliverdin is not native to E. coli, the addition of heme oxygenase is required for the synthesis of bilivedin, enabling the self-assembly of the light switch. In 2011, the Macquarie Team successfully managed to construct and characterise the heme oxygenase 1 as a BioBrick. They showed, via its green color, that cells expressing the heme oxygenase could degrade heme into biliverdin.
In this project we will aim to construct the BioBricks using Gibson cloning as opposed to restriction enzyme digests.
Project Work Flow

 "
"