Team:MIT/Results
From 2012.igem.org
(Difference between revisions)
| Line 124: | Line 124: | ||
<br> | <br> | ||
| + | </div> | ||
| + | |||
| + | <div class="bio" id="b3bio"> | ||
| + | |||
| + | <b>In Vivo RNA Strand Displacement </b> | ||
| + | <br> | ||
| + | <br> | ||
| + | <b> Strategy 1: Take DNA sequences that worked in In Vitro Strand Displacement, convert to RNA, transfect inside HEK293 cells </b> | ||
| + | <br> | ||
| + | <br> | ||
| + | <b> Strategy 2: Tag RNA strand with an Alexa Fluorophore to act as a transfection marker </b> | ||
| + | <br> | ||
| + | <br> | ||
| + | <b> Strategy 3: Create DNA plasmids driving transcription of RNA inputs, while transfecting RNA Reporter </b> | ||
| + | <br> | ||
| + | <br> | ||
| + | <b> Strategy 4: Nucleofect RNA reporter, RNA inputs </b> | ||
| + | <br> | ||
| + | <br> | ||
| + | <b> [[Strategy 5]]: Redesign RNA Reporter </b> | ||
</div> | </div> | ||
Revision as of 04:23, 25 September 2012
RNA Strand Displacement In Vitro
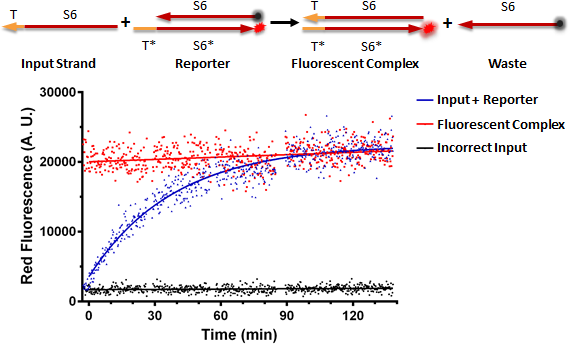

Nucleic Acid Delivery
In Vivo RNA Strand Displacement
Strategy 1: Take DNA sequences that worked in In Vitro Strand Displacement, convert to RNA, transfect inside HEK293 cells
Strategy 2: Tag RNA strand with an Alexa Fluorophore to act as a transfection marker
Strategy 3: Create DNA plasmids driving transcription of RNA inputs, while transfecting RNA Reporter
Strategy 4: Nucleofect RNA reporter, RNA inputs
[[Strategy 5]]: Redesign RNA Reporter
Strategy 1: Take DNA sequences that worked in In Vitro Strand Displacement, convert to RNA, transfect inside HEK293 cells
Strategy 2: Tag RNA strand with an Alexa Fluorophore to act as a transfection marker
Strategy 3: Create DNA plasmids driving transcription of RNA inputs, while transfecting RNA Reporter
Strategy 4: Nucleofect RNA reporter, RNA inputs
[[Strategy 5]]: Redesign RNA Reporter
 "
"

